Correlation between Cancer Stem Cells, Inflammation and Malignant Transformation in a DEN-Induced Model of Hepatic Carcinogenesis
- PMID: 35877422
- PMCID: PMC9324326
- DOI: 10.3390/cimb44070198
Correlation between Cancer Stem Cells, Inflammation and Malignant Transformation in a DEN-Induced Model of Hepatic Carcinogenesis
Abstract
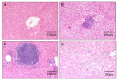
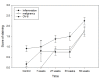
Chronic inflammation and cancer stem cells are known risk factors for tumorigenesis. The aetiology of hepatocellular carcinoma (HCC) involves a multistep pathological process that is characterised by chronic inflammation and hepatocyte damage, but the correlation between HCC, inflammation and cancer stem cells remains unclear. In this study, we examined the role of hepatic progenitor cells in a mouse model of chemical-induced hepatocarcinogenesis to elucidate the relationship between inflammation, malignant transformation and cancer stem cells. We used diethylnitrosamine (DEN) to induce liver tumour and scored for H&E and reticulin staining. We also scored for immunohistochemistry staining for OV-6 expression and analysed the statistical correlation between them. DEN progressively induced inflammation at week 7 (40%, 2/5); week 27 (75%, 6/8); week 33 (62.5%, 5/8); and week 50 (100%, 12/12). DEN progressively induced malignant transformation at week 7 (0%, 0/5); week 27 (87.5%, 7/8); week 33 (100%, 8/8); and week 50 (100%, 12/12). The obtained data showed that DEN progressively induced high-levels of OV-6 expression at week 7 (20%, 1/5); week 27 (37.5%, 3/8); week 33 (50%, 4/8); and week 50 (100%, 12/12). DEN-induced inflammation, malignant transformation and high-level OV-6 expression in hamster liver, as shown above, as well as applying Spearman’s correlation to the data showed that the expression of OV-6 was significantly correlated to inflammation (p = 0.001) and malignant transformation (p < 0.001). There was a significant correlation between the number of cancer stem cells, inflammation and malignant transformation in a DEN-induced model of hepatic carcinogenesis in the hamster.
Keywords: diethylnitrosamine; hepatocellular carcinoma; inflammation.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


Similar articles
-
Inhibition of androgen/AR signaling inhibits diethylnitrosamine (DEN) induced tumour initiation and remodels liver immune cell networks.Sci Rep. 2021 Feb 11;11(1):3646. doi: 10.1038/s41598-021-82252-x. Sci Rep. 2021. PMID: 33574348 Free PMC article.
-
High-saturate-fat diet delays initiation of diethylnitrosamine-induced hepatocellular carcinoma.BMC Gastroenterol. 2014 Nov 20;14:195. doi: 10.1186/s12876-014-0195-9. BMC Gastroenterol. 2014. PMID: 25410681 Free PMC article.
-
Chronic administration of diethylnitrosamine to induce hepatocarcinogenesis and to evaluate its synergistic effect with other hepatotoxins in mice.Toxicol Appl Pharmacol. 2019 Sep 1;378:114611. doi: 10.1016/j.taap.2019.114611. Epub 2019 Jun 6. Toxicol Appl Pharmacol. 2019. PMID: 31176654
-
Diethylnitrosamine-induced liver tumorigenesis in mice.Methods Cell Biol. 2021;163:137-152. doi: 10.1016/bs.mcb.2020.08.006. Epub 2020 Oct 2. Methods Cell Biol. 2021. PMID: 33785162 Review.
-
Correlation of O4-ethyldeoxythymidine accumulation, hepatic initiation and hepatocellular carcinoma induction in rats continuously administered diethylnitrosamine.Carcinogenesis. 1986 Feb;7(2):241-6. doi: 10.1093/carcin/7.2.241. Carcinogenesis. 1986. PMID: 2868805
Cited by
-
The role of lipophagy in liver cancer: mechanisms and targeted therapeutic interventions.Front Cell Dev Biol. 2025 Jul 8;13:1562542. doi: 10.3389/fcell.2025.1562542. eCollection 2025. Front Cell Dev Biol. 2025. PMID: 40698038 Free PMC article. Review.
References
-
- Beier D., Hau P., Proescholdt M., Lohmeier A., Wischhusen J., Oefner P.J., Aigner L., Brawanski A., Bogdahn U., Beier C.P. CD133(+) and CD133(−) glioblastoma-derived cancer stem cells show differential growth characteristics and molecular profiles. Cancer Res. 2007;67:4010–4015. doi: 10.1158/0008-5472.CAN-06-4180. - DOI - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources

