Systematic whole-genome sequencing reveals an unexpected diversity among actinomycetoma pathogens and provides insights into their antibacterial susceptibilities
- PMID: 35877680
- PMCID: PMC9352199
- DOI: 10.1371/journal.pntd.0010128
Systematic whole-genome sequencing reveals an unexpected diversity among actinomycetoma pathogens and provides insights into their antibacterial susceptibilities
Abstract
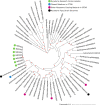
Mycetoma is a neglected tropical chronic granulomatous inflammatory disease of the skin and subcutaneous tissues. More than 70 species with a broad taxonomic diversity have been implicated as agents of mycetoma. Understanding the full range of causative organisms and their antibiotic sensitivity profiles are essential for the appropriate treatment of infections. The present study focuses on the analysis of full genome sequences and antibiotic inhibitory concentration profiles of actinomycetoma strains from patients seen at the Mycetoma Research Centre in Sudan with a view to developing rapid diagnostic tests. Seventeen pathogenic isolates obtained by surgical biopsies were sequenced using MinION and Illumina methods, and their antibiotic inhibitory concentration profiles determined. The results highlight an unexpected diversity of actinomycetoma causing pathogens, including three Streptomyces isolates assigned to species not previously associated with human actinomycetoma and one new Streptomyces species. Thus, current approaches for clinical and histopathological classification of mycetoma may need to be updated. The standard treatment for actinomycetoma is a combination of sulfamethoxazole/trimethoprim and amoxicillin/clavulanic acid. Most tested isolates had a high IC (inhibitory concentration) to sulfamethoxazole/trimethoprim or to amoxicillin alone. However, the addition of the β-lactamase inhibitor clavulanic acid to amoxicillin increased susceptibility, particularly for Streptomyces somaliensis and Streptomyces sudanensis. Actinomadura madurae isolates appear to have a particularly high IC under laboratory conditions, suggesting that alternative agents, such as amikacin, could be considered for more effective treatment. The results obtained will inform future diagnostic methods for the identification of actinomycetoma and treatment.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures






References
-
- Hay RJ, Asiedu KB, Fahal AH. Mycetoma–a long journey out of the shadows. Trans R Soc Trop Med Hyg. 2020;115: 281–282. - PubMed
-
- WHO. Neglected tropical diseases. [cited 8 Feb 2016]. Available: http://www.who.int/neglected_diseases/diseases/en/
Publication types
MeSH terms
Substances
Associated data
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

