Rational Discovery of Antimicrobial Peptides by Means of Artificial Intelligence
- PMID: 35877911
- PMCID: PMC9320227
- DOI: 10.3390/membranes12070708
Rational Discovery of Antimicrobial Peptides by Means of Artificial Intelligence
Abstract
Antibiotic resistance is a worldwide public health problem due to the costs and mortality rates it generates. However, the large pharmaceutical industries have stopped searching for new antibiotics because of their low profitability, given the rapid replacement rates imposed by the increasingly observed resistance acquired by microorganisms. Alternatively, antimicrobial peptides (AMPs) have emerged as potent molecules with a much lower rate of resistance generation. The discovery of these peptides is carried out through extensive in vitro screenings of either rational or non-rational libraries. These processes are tedious and expensive and generate only a few AMP candidates, most of which fail to show the required activity and physicochemical properties for practical applications. This work proposes implementing an artificial intelligence algorithm to reduce the required experimentation and increase the efficiency of high-activity AMP discovery. Our deep learning (DL) model, called AMPs-Net, outperforms the state-of-the-art method by 8.8% in average precision. Furthermore, it is highly accurate to predict the antibacterial and antiviral capacity of a large number of AMPs. Our search led to identifying two unreported antimicrobial motifs and two novel antimicrobial peptides related to them. Moreover, by coupling DL with molecular dynamics (MD) simulations, we were able to find a multifunctional peptide with promising therapeutic effects. Our work validates our previously proposed pipeline for a more efficient rational discovery of novel AMPs.
Keywords: antimicrobial; artificial intelligence; graphs; molecular dynamics; peptides.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
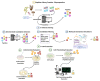
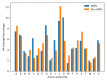
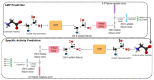
Similar articles
-
Design, Screening, and Testing of Non-Rational Peptide Libraries with Antimicrobial Activity: In Silico and Experimental Approaches.Antibiotics (Basel). 2020 Nov 30;9(12):854. doi: 10.3390/antibiotics9120854. Antibiotics (Basel). 2020. PMID: 33265897 Free PMC article. Review.
-
Combining genetic algorithm with machine learning strategies for designing potent antimicrobial peptides.BMC Bioinformatics. 2021 May 11;22(1):239. doi: 10.1186/s12859-021-04156-x. BMC Bioinformatics. 2021. PMID: 33975547 Free PMC article.
-
AI4AMP: an Antimicrobial Peptide Predictor Using Physicochemical Property-Based Encoding Method and Deep Learning.mSystems. 2021 Dec 21;6(6):e0029921. doi: 10.1128/mSystems.00299-21. Epub 2021 Nov 16. mSystems. 2021. PMID: 34783578 Free PMC article.
-
CalcAMP: A New Machine Learning Model for the Accurate Prediction of Antimicrobial Activity of Peptides.Antibiotics (Basel). 2023 Apr 7;12(4):725. doi: 10.3390/antibiotics12040725. Antibiotics (Basel). 2023. PMID: 37107088 Free PMC article.
-
Innovative Strategies and Methodologies in Antimicrobial Peptide Design.J Funct Biomater. 2024 Oct 29;15(11):320. doi: 10.3390/jfb15110320. J Funct Biomater. 2024. PMID: 39590524 Free PMC article. Review.
Cited by
-
Heterologous Production of Antimicrobial Peptides: Notes to Consider.Protein J. 2024 Apr;43(2):129-158. doi: 10.1007/s10930-023-10174-w. Epub 2024 Jan 5. Protein J. 2024. PMID: 38180586
-
Evaluating the impact of cell-penetrating motif position on the cellular uptake of magnetite nanoparticles.Front Bioeng Biotechnol. 2024 Dec 2;12:1450694. doi: 10.3389/fbioe.2024.1450694. eCollection 2024. Front Bioeng Biotechnol. 2024. PMID: 39687269 Free PMC article.
-
Recent Progress in the Discovery and Design of Antimicrobial Peptides Using Traditional Machine Learning and Deep Learning.Antibiotics (Basel). 2022 Oct 21;11(10):1451. doi: 10.3390/antibiotics11101451. Antibiotics (Basel). 2022. PMID: 36290108 Free PMC article. Review.
-
Biopreservation strategies using bacteriocins to control meat spoilage and foodborne outbreaks.Ital J Food Saf. 2024 Oct 17;13(4):12558. doi: 10.4081/ijfs.2024.12558. eCollection 2024 Nov 12. Ital J Food Saf. 2024. PMID: 39749182 Free PMC article. Review.
-
AbAMPdb: a database of Acinetobacter baumannii specific antimicrobial peptides.Database (Oxford). 2024 Oct 12;2024:baae096. doi: 10.1093/database/baae096. Database (Oxford). 2024. PMID: 39395188 Free PMC article.
References
-
- Stokowski L.A. Antimicrobial Resistance: A Primer. 2010. [(accessed on 15 November 2021)]. Available online: https://www.medscape.com/viewarticle/729196_2.
-
- Centers for Disease Control and Prevention About Antibiotic Resistance. [(accessed on 15 November 2021)];2019 Available online: https://www.cdc.gov/drugresistance/about.html.
Grants and funding
LinkOut - more resources
Full Text Sources

