Protons in Gating the Kv1.2 Channel: A Calculated Set of Protonation States in Response to Polarization/Depolarization of the Channel, with the Complete Proposed Proton Path from Voltage Sensing Domain to Gate
- PMID: 35877921
- PMCID: PMC9318985
- DOI: 10.3390/membranes12070718
Protons in Gating the Kv1.2 Channel: A Calculated Set of Protonation States in Response to Polarization/Depolarization of the Channel, with the Complete Proposed Proton Path from Voltage Sensing Domain to Gate
Abstract
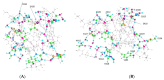
We have in the past proposed that proton motion constitutes the gating current in the potassium channel Kv1.2 and is responsible for the gating mechanism. For this to happen, there must be a proton path between the voltage-sensing domain (VSD) and the channel gate, and here we present quantum calculations that lead to a specific pair of proton paths, defined at the molecular level, with well-defined water molecule linkages, and with hydrogen bonding between residues; there is also at least one interpath crossover, where protons can switch paths. Quantum calculations on the entire 563-atom system give the complete geometry, the energy, and atomic charges. Calculations show that three specific residues (in the pdb 3Lut numbering, H418, E327, R326), and the T1 intracellular moiety, all of which have been shown experimentally to be involved in gating, would necessarily be protonated or deprotonated in the path between the VSD and the gate. Hydroxyl reorientation of serine and threonine residues are shown to provide a means of adjusting proton directions of motion. In the deprotonated state for K312, a low energy state, our calculations come close to reproducing the X-ray structure. The demonstration of the existence of a double proton path between VSD and gate supports the proposed proton gating mechanism; when combined with our earlier demonstration of proton generation in the VSD, and comparison with other systems that are known to move protons, we are close to achieving the definition of a complete gating mechanism in molecular detail. The coupling of the paths to the VSD, and to the PVPV section that essentially forms the gate, can be easily seen from the results of the calculation. The gate itself remains for further computations.
Keywords: amino acid strings; ion channel gating; proton transport paths.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

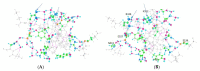


Similar articles
-
Water, Protons, and the Gating of Voltage-Gated Potassium Channels.Membranes (Basel). 2024 Jan 29;14(2):37. doi: 10.3390/membranes14020037. Membranes (Basel). 2024. PMID: 38392664 Free PMC article. Review.
-
The Role of Proton Transport in Gating Current in a Voltage Gated Ion Channel, as Shown by Quantum Calculations.Sensors (Basel). 2018 Sep 18;18(9):3143. doi: 10.3390/s18093143. Sensors (Basel). 2018. PMID: 30231473 Free PMC article.
-
Quantum Calculation of Proton and Other Charge Transfer Steps in Voltage Sensing in the Kv1.2 Channel.J Phys Chem B. 2019 Sep 26;123(38):7984-7998. doi: 10.1021/acs.jpcb.9b05448. Epub 2019 Sep 17. J Phys Chem B. 2019. PMID: 31441655
-
Structural and molecular insight into the pH-induced low-permeability of the voltage-gated potassium channel Kv1.2 through dewetting of the water cavity.PLoS Comput Biol. 2020 Apr 21;16(4):e1007405. doi: 10.1371/journal.pcbi.1007405. eCollection 2020 Apr. PLoS Comput Biol. 2020. PMID: 32315300 Free PMC article.
-
Voltage gated ion channel function: gating, conduction, and the role of water and protons.Int J Mol Sci. 2012;13(2):1680-1709. doi: 10.3390/ijms13021680. Epub 2012 Feb 6. Int J Mol Sci. 2012. PMID: 22408417 Free PMC article. Review.
Cited by
-
Water, Protons, and the Gating of Voltage-Gated Potassium Channels.Membranes (Basel). 2024 Jan 29;14(2):37. doi: 10.3390/membranes14020037. Membranes (Basel). 2024. PMID: 38392664 Free PMC article. Review.
-
H+ and Confined Water in Gating in Many Voltage-Gated Potassium Channels: Ion/Water/Counterion/Protein Networks and Protons Added to Gate the Channel.Int J Mol Sci. 2025 Jul 29;26(15):7325. doi: 10.3390/ijms26157325. Int J Mol Sci. 2025. PMID: 40806455 Free PMC article. Review.
References
-
- Hille B. Ion Channels of Excitable Membranes. 3rd ed. Sinauer Associates; Sunderland, MA, USA: 2001.
-
- Vargas E., Yarov-Yarovoy V., Khalili-Araghi F., Catterall W.A., Klein M.L., Tarek M., Lindahl E., Schulten K., Perozo E., Bezanilla F., et al. An emerging consensus on voltage-dependent gating from computational modeling and molecular dynamics simulations. J. Gen. Physiol. 2012;140:587–594. doi: 10.1085/jgp.201210873. - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Miscellaneous

