Exploring the Utility of ssDNA Aptamers Directed against Snake Venom Toxins as New Therapeutics for Snakebite Envenoming
- PMID: 35878207
- PMCID: PMC9318713
- DOI: 10.3390/toxins14070469
Exploring the Utility of ssDNA Aptamers Directed against Snake Venom Toxins as New Therapeutics for Snakebite Envenoming
Abstract
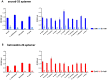
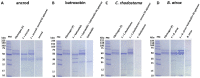
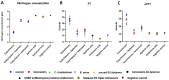
Snakebite is a neglected tropical disease that causes considerable death and disability in the tropical world. Although snakebite can cause a variety of pathologies in victims, haemotoxic effects are particularly common and are typically characterised by haemorrhage and/or venom-induced consumption coagulopathy. Antivenoms are the mainstay therapy for treating the toxic effects of snakebite, but despite saving thousands of lives annually, these therapies are associated with limited cross-snake species efficacy due to venom variation, which ultimately restricts their therapeutic utility to particular geographical regions. In this study, we sought to explore the potential of ssDNA aptamers as toxin-specific inhibitory alternatives to antibodies. As a proof of principle model, we selected snake venom serine protease toxins, which are responsible for contributing to venom-induced coagulopathy following snakebite envenoming, as our target. Using SELEX technology, we selected ssDNA aptamers against recombinantly expressed versions of the fibrinogenolytic SVSPs ancrod from the venom of C. rhodostoma and batroxobin from B. atrox. From the resulting pool of specific ssDNA aptamers directed against each target, we identified candidates that exhibited low nanomolar binding affinities to their targets. Downstream aptamer-linked immobilised sorbent assay, fibrinogenolysis, and coagulation profiling experiments demonstrated that the candidate aptamers were able to recognise native and recombinant SVSP toxins and inhibit the toxin- and venom-induced prolongation of plasma clotting times and the consumption of fibrinogen, with inhibitory potencies highly comparable to commercial polyvalent antivenoms. Our findings demonstrate that rationally selected toxin-specific aptamers can exhibit broad in vitro cross-reactivity against toxin isoforms found in different snake venoms and are capable of inhibiting toxins in pathologically relevant in vitro and ex vivo models of venom activity. These data highlight the potential utility of ssDNA aptamers as novel toxin-inhibiting therapeutics of value for tackling snakebite envenoming.
Keywords: SELEX selection; in vitro assays; recombinant toxins; snake venom serine proteases; snakebite envenoming; ssDNA aptamers.
Conflict of interest statement
The authors declare no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Figures


Similar articles
-
Exploring the Utility of Recombinant Snake Venom Serine Protease Toxins as Immunogens for Generating Experimental Snakebite Antivenoms.Toxins (Basel). 2022 Jun 29;14(7):443. doi: 10.3390/toxins14070443. Toxins (Basel). 2022. PMID: 35878181 Free PMC article.
-
Pathology-specific experimental antivenoms for haemotoxic snakebite: The impact of immunogen diversity on the in vitro cross-reactivity and in vivo neutralisation of geographically diverse snake venoms.PLoS Negl Trop Dis. 2021 Aug 18;15(8):e0009659. doi: 10.1371/journal.pntd.0009659. eCollection 2021 Aug. PLoS Negl Trop Dis. 2021. PMID: 34407084 Free PMC article.
-
A Review of the Proteomic Profiling of African Viperidae and Elapidae Snake Venoms and Their Antivenom Neutralisation.Toxins (Basel). 2022 Oct 22;14(11):723. doi: 10.3390/toxins14110723. Toxins (Basel). 2022. PMID: 36355973 Free PMC article. Review.
-
Haemotoxic snake venoms: their functional activity, impact on snakebite victims and pharmaceutical promise.Br J Haematol. 2017 Jun;177(6):947-959. doi: 10.1111/bjh.14591. Epub 2017 Feb 24. Br J Haematol. 2017. PMID: 28233897 Free PMC article. Review.
-
Commercial Antivenoms Exert Broad Paraspecific Immunological Binding and In Vitro Inhibition of Medically Important Bothrops Pit Viper Venoms.Toxins (Basel). 2022 Dec 20;15(1):1. doi: 10.3390/toxins15010001. Toxins (Basel). 2022. PMID: 36668821 Free PMC article.
Cited by
-
De novo designed proteins neutralize lethal snake venom toxins.Nature. 2025 Mar;639(8053):225-231. doi: 10.1038/s41586-024-08393-x. Epub 2025 Jan 15. Nature. 2025. PMID: 39814879 Free PMC article.
-
Considerations for the development of a field-based medical device for the administration of adjunctive therapies for snakebite envenoming.Toxicon X. 2023 Aug 19;20:100169. doi: 10.1016/j.toxcx.2023.100169. eCollection 2023 Dec. Toxicon X. 2023. PMID: 37661997 Free PMC article.
-
Low-Cost Point-of-Care Monitoring of ALT and AST Is Promising for Faster Decision Making and Diagnosis of Acute Liver Injury.Diagnostics (Basel). 2023 Sep 16;13(18):2967. doi: 10.3390/diagnostics13182967. Diagnostics (Basel). 2023. PMID: 37761334 Free PMC article.
-
Advanced Situation with Recombinant Toxins: Diversity, Production and Application Purposes.Int J Mol Sci. 2023 Feb 27;24(5):4630. doi: 10.3390/ijms24054630. Int J Mol Sci. 2023. PMID: 36902061 Free PMC article. Review.
-
Towards better antivenoms: navigating the road to new types of snakebite envenoming therapies.J Venom Anim Toxins Incl Trop Dis. 2023 Dec 18;29:e20230057. doi: 10.1590/1678-9199-JVATITD-2023-0057. eCollection 2023. J Venom Anim Toxins Incl Trop Dis. 2023. PMID: 38116472 Free PMC article.
References
-
- Kasturiratne A., Wickremasinghe A.R., de Silva N., Gunawardena N.K., Pathmeswaran A., Premaratna R., Savioli L., Lalloo D.G., de Silva H.J. The global burden of snakebite: A literature analysis and modelling based on regional estimates of envenoming and deaths. PLoS Med. 2008;5:e218. doi: 10.1371/journal.pmed.0050218. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

