The dark septate endophyte Phialocephala sphaeroides suppresses conifer pathogen transcripts and promotes root growth of Norway spruce
- PMID: 35878416
- PMCID: PMC9743008
- DOI: 10.1093/treephys/tpac089
The dark septate endophyte Phialocephala sphaeroides suppresses conifer pathogen transcripts and promotes root growth of Norway spruce
Abstract
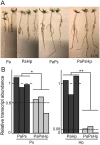
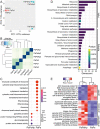
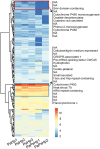
Plant-associated microbes including dark septate endophytes (DSEs) of forest trees play diverse functional roles in host fitness including growth promotion and increased defence. However, little is known about the impact on the fungal transcriptome and metabolites during tripartite interaction involving plant host, endophyte and pathogen. To understand the transcriptional regulation of endophyte and pathogen during co-infection, Norway spruce (Picea abies) seedlings were infected with DSE Phialocephala sphaeroides, or conifer root-rot pathogen Heterobasidion parviporum, or both. Phialocephala sphaeroides showed low but stable transcripts abundance (a decrease of 40%) during interaction with Norway spruce and conifer pathogen. By contrast, H. parviporum transcripts were significantly reduced (92%) during co-infection. With RNA sequencing analysis, P. sphaeroides experienced a shift from cell growth to anti-stress and antagonistic responses, while it repressed the ability of H. parviporum to access carbohydrate nutrients by suppressing its carbohydrate/polysaccharide-degrading enzyme machinery. The pathogen on the other hand secreted cysteine peptidase to restrict free growth of P. sphaeroides. The expression of both DSE P. sphaeroides and pathogen H. parviporum genes encoding plant growth promotion products were equally detected in both dual and tripartite interaction systems. This was further supported by the presence of tryptophan-dependent indolic compound in liquid culture of P. sphaeroides. Norway spruce and Arabidopsis seedlings treated with P. sphaeroides culture filtrate exhibited auxin-like phenotypes, such as enhanced root hairs, and primary root elongation at low concentration but shortened primary root at high concentration. The results suggested that the presence of the endophyte had strong repressive or suppressive effect on H. parviporum transcripts encoding genes involved in nutrient acquisition.
Keywords: dark septate endophyte; fungi–fungi–plant interaction; growth promotion; transcriptome.
© The Author(s) 2022. Published by Oxford University Press.
Figures


Similar articles
-
The dark septate endophyte Phialocephala sphaeroides confers growth fitness benefits and mitigates pathogenic effects of Heterobasidion on Norway spruce.Tree Physiol. 2022 Apr 7;42(4):891-906. doi: 10.1093/treephys/tpab147. Tree Physiol. 2022. PMID: 34791486 Free PMC article.
-
Molecular and Chemical Screening for Inherent Disease Resistance Factors of Norway Spruce (Picea abies) Clones Against Conifer Stem Rot Pathogen Heterobasidion parviporum.Phytopathology. 2022 Apr;112(4):872-880. doi: 10.1094/PHYTO-09-21-0379-R. Epub 2022 Mar 18. Phytopathology. 2022. PMID: 34698543
-
The pathogenic white-rot fungus Heterobasidion parviporum triggers non-specific defence responses in the bark of Norway spruce.Tree Physiol. 2011 Nov;31(11):1262-72. doi: 10.1093/treephys/tpr113. Tree Physiol. 2011. PMID: 22084022
-
Dual RNA-seq analysis provides new insights into interactions between Norway spruce and necrotrophic pathogen Heterobasidion annosum s.l.BMC Plant Biol. 2019 Jan 3;19(1):2. doi: 10.1186/s12870-018-1602-0. BMC Plant Biol. 2019. PMID: 30606115 Free PMC article.
-
The Conifer Root and Stem Rot Pathogen (Heterobasidion parviporum): Effectome Analysis and Roles in Interspecific Fungal Interactions.Microorganisms. 2019 Dec 5;7(12):658. doi: 10.3390/microorganisms7120658. Microorganisms. 2019. PMID: 31817407 Free PMC article.
Cited by
-
The Diverse Mycorrizal Morphology of Rhododendron dauricum, the Fungal Communities Structure and Dynamics from the Mycorrhizosphere.J Fungi (Basel). 2024 Jan 14;10(1):65. doi: 10.3390/jof10010065. J Fungi (Basel). 2024. PMID: 38248974 Free PMC article.
-
The morphoanatomy of Serjania erecta Radlk (Sapindaceae) provides evidence of biotrophic interactions by endophytic fungi within leaves.PeerJ. 2023 Sep 15;11:e15980. doi: 10.7717/peerj.15980. eCollection 2023. PeerJ. 2023. PMID: 37727689 Free PMC article.
-
The Three-Dimensional Structure of the Genome of the Dark Septate Endophyte Exophiala tremulae and Its Symbiosis Effect on Alpine Meadow Plant Growth.J Fungi (Basel). 2025 Mar 24;11(4):246. doi: 10.3390/jof11040246. J Fungi (Basel). 2025. PMID: 40278067 Free PMC article.
-
The temperate forest phyllosphere and rhizosphere microbiome: a case study of sugar maple.Front Microbiol. 2025 Jan 15;15:1504444. doi: 10.3389/fmicb.2024.1504444. eCollection 2024. Front Microbiol. 2025. PMID: 39881993 Free PMC article. Review.
-
Beyond the surface: exploring the mycobiome of Norway spruce under drought stress and with Heterobasidion parviporum.BMC Microbiol. 2023 Nov 17;23(1):350. doi: 10.1186/s12866-023-03099-y. BMC Microbiol. 2023. PMID: 37978432 Free PMC article.
References
-
- Afzal I, Iqrar I, Shinwari ZK, Yasmin A (2017) Plant growth-promoting potential of endophytic bacteria isolated from roots of wild Dodonaea viscosa L. Plant Growth Regul 81:399–408.
-
- Alberton O, Kuyper TW, Summerbell RC (2010) Dark septate root endophytic fungi increase growth of Scots pine seedlings under elevated CO2 through enhanced nitrogen use efficiency. Plant Soil 328:459–470.
-
- Berglund M, Rönnberg J (2004) Effectiveness of treatment of Norway spruce stumps with Phlebiopsis gigantea at different rates of coverage for the control of Heterobasidion. For Pathol 34:233–243.

