Phenylalanine impairs insulin signaling and inhibits glucose uptake through modification of IRβ
- PMID: 35879296
- PMCID: PMC9314339
- DOI: 10.1038/s41467-022-32000-0
Phenylalanine impairs insulin signaling and inhibits glucose uptake through modification of IRβ
Abstract
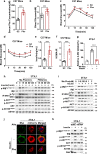
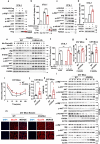
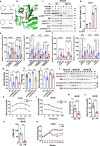
Whether amino acids act on cellular insulin signaling remains unclear, given that increased circulating amino acid levels are associated with the onset of type 2 diabetes (T2D). Here, we report that phenylalanine modifies insulin receptor beta (IRβ) and inactivates insulin signaling and glucose uptake. Mice fed phenylalanine-rich chow or phenylalanine-producing aspartame or overexpressing human phenylalanyl-tRNA synthetase (hFARS) develop insulin resistance and T2D symptoms. Mechanistically, FARS phenylalanylate lysine 1057/1079 of IRβ (F-K1057/1079), inactivating IRβ and preventing insulin from promoting glucose uptake by cells. SIRT1 reverse F-K1057/1079 and counteract the insulin-inactivating effects of hFARS and phenylalanine. F-K1057/1079 and SIRT1 levels in white blood cells from T2D patients are positively and negatively correlated with T2D onset, respectively. Blocking F-K1057/1079 with phenylalaninol sensitizes insulin signaling and relieves T2D symptoms in hFARS-transgenic and db/db mice. These findings shed light on the activation of insulin signaling and T2D progression through inhibition of phenylalanylation.
© 2022. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures




References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Miscellaneous

