Circular RNA ROCK1, a novel circRNA, suppresses osteosarcoma proliferation and migration via altering the miR-532-5p/PTEN axis
- PMID: 35879346
- PMCID: PMC9356001
- DOI: 10.1038/s12276-022-00806-z
Circular RNA ROCK1, a novel circRNA, suppresses osteosarcoma proliferation and migration via altering the miR-532-5p/PTEN axis
Abstract
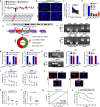
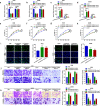
As the most prevalent bone tumor in children and adolescents, the pathogenesis and metastasis of osteosarcoma (OS) remain largely unclear. Here, we investigated the expression and function of a novel circular RNA (circRNA), circROCK1-E3/E4, which is back-spliced from exons 3 and 4 of Rho-associated coiled-coil containing protein kinase 1 (ROCK1) in OS. We found that circROCK1-E3/E4, regulated by the well-known RNA-binding protein quaking (QKI), was downregulated in OS and correlated with unfavorable clinical features of patients with OS. Functional proliferation and cell motility assays indicated that circROCK1-E3/E4 serves as a tumor suppressor in OS cells. Mechanistically, circROCK1-E3/E4 suppressed proliferation and migration by upregulating phosphatase and tensin homolog (PTEN) through microRNA-532-5p (miR-532-5p) sponging. In the constructed nude mouse model, circROCK1-E3/E4 inhibited tumor growth and lung metastasis in vivo. This study demonstrates the functions and molecular mechanisms of circROCK1-E3/E4 in the progression of OS. These findings may identify novel targets for the molecular therapy of OS.
© 2022. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures





Similar articles
-
Long noncoding RNA DANCR, working as a competitive endogenous RNA, promotes ROCK1-mediated proliferation and metastasis via decoying of miR-335-5p and miR-1972 in osteosarcoma.Mol Cancer. 2018 May 12;17(1):89. doi: 10.1186/s12943-018-0837-6. Mol Cancer. 2018. PMID: 29753317 Free PMC article.
-
MicroRNA-129-5p suppresses cell proliferation, migration and invasion via targeting ROCK1 in osteosarcoma.Mol Med Rep. 2018 Mar;17(3):4777-4784. doi: 10.3892/mmr.2018.8374. Epub 2018 Jan 4. Mol Med Rep. 2018. PMID: 29328417
-
Circ_0001174 facilitates osteosarcoma cell proliferation, migration, and invasion by targeting the miR-186-5p/MACC1 axis.J Orthop Surg Res. 2022 Mar 12;17(1):159. doi: 10.1186/s13018-022-03059-8. J Orthop Surg Res. 2022. PMID: 35279159 Free PMC article.
-
circUSP34 accelerates osteosarcoma malignant progression by sponging miR-16-5p.Cancer Sci. 2022 Jan;113(1):120-131. doi: 10.1111/cas.15147. Epub 2021 Nov 22. Cancer Sci. 2022. PMID: 34592064 Free PMC article.
-
miR-139 inhibits osteosarcoma cell proliferation and invasion by targeting ROCK1.Front Biosci (Landmark Ed). 2019 Jun 1;24(6):1167-1177. doi: 10.2741/4773. Front Biosci (Landmark Ed). 2019. PMID: 31136973
Cited by
-
CNTNAP4 signaling regulates osteosarcoma disease progression.NPJ Precis Oncol. 2023 Jan 4;7(1):2. doi: 10.1038/s41698-022-00344-x. NPJ Precis Oncol. 2023. PMID: 36599925 Free PMC article.
-
DCC-2036 inhibits osteosarcoma via targeting HCK and the PI3K/AKT-mTORC1 axis to promote autophagy.World J Surg Oncol. 2025 Apr 2;23(1):115. doi: 10.1186/s12957-025-03778-2. World J Surg Oncol. 2025. PMID: 40176057 Free PMC article.
-
Functional role of MicroRNA/PI3K/AKT axis in osteosarcoma.Front Oncol. 2023 Jun 19;13:1219211. doi: 10.3389/fonc.2023.1219211. eCollection 2023. Front Oncol. 2023. PMID: 37404761 Free PMC article. Review.
-
Hsa_circ_0001304 promotes vascular neointimal hyperplasia accompanied by autophagy activation.Commun Biol. 2025 Jan 30;8(1):146. doi: 10.1038/s42003-025-07580-4. Commun Biol. 2025. PMID: 39881153 Free PMC article.
-
Circ_C4orf36 Promotes the Proliferation and Osteogenic Differentiation of BMSCs by Regulating VEGFA.Biochem Genet. 2023 Jun;61(3):931-944. doi: 10.1007/s10528-022-10290-9. Epub 2022 Oct 15. Biochem Genet. 2023. PMID: 36242722
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials

