Uncovering the mode of action of engineered T cells in patient cancer organoids
- PMID: 35879361
- PMCID: PMC9849137
- DOI: 10.1038/s41587-022-01397-w
Uncovering the mode of action of engineered T cells in patient cancer organoids
Abstract
Extending the success of cellular immunotherapies against blood cancers to the realm of solid tumors will require improved in vitro models that reveal therapeutic modes of action at the molecular level. Here we describe a system, called BEHAV3D, developed to study the dynamic interactions of immune cells and patient cancer organoids by means of imaging and transcriptomics. We apply BEHAV3D to live-track >150,000 engineered T cells cultured with patient-derived, solid-tumor organoids, identifying a 'super engager' behavioral cluster comprising T cells with potent serial killing capacity. Among other T cell concepts we also study cancer metabolome-sensing engineered T cells (TEGs) and detect behavior-specific gene signatures that include a group of 27 genes with no previously described T cell function that are expressed by super engager killer TEGs. We further show that type I interferon can prime resistant organoids for TEG-mediated killing. BEHAV3D is a promising tool for the characterization of behavioral-phenotypic heterogeneity of cellular immunotherapies and may support the optimization of personalized solid-tumor-targeting cell therapies.
© 2022. The Author(s).
Conflict of interest statement
H.C., Y.E.B.-E., K.K. and J.F.D. are named as inventors on (pending) patents related to the organoid technology. For the full disclosure of H.C., see
Figures

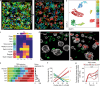




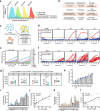
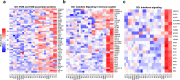



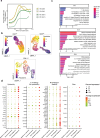
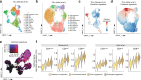
Comment in
-
BEHAV3D: an imaging and transcriptomics platform that unravels T cell antitumor activity.Nat Biotechnol. 2023 Jan;41(1):31-32. doi: 10.1038/s41587-022-01398-9. Nat Biotechnol. 2023. PMID: 35902779 No abstract available.
References
-
- Chen, D. S. & Mellman, I. Elements of cancer immunity and the cancer-immune set point. Nature541, 321–330 (2017). - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

