Therapeutic targeting of ATR in alveolar rhabdomyosarcoma
- PMID: 35879366
- PMCID: PMC9314382
- DOI: 10.1038/s41467-022-32023-7
Therapeutic targeting of ATR in alveolar rhabdomyosarcoma
Abstract
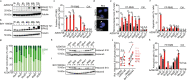
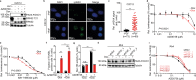
Despite advances in multi-modal treatment approaches, clinical outcomes of patients suffering from PAX3-FOXO1 fusion oncogene-expressing alveolar rhabdomyosarcoma (ARMS) remain dismal. Here we show that PAX3-FOXO1-expressing ARMS cells are sensitive to pharmacological ataxia telangiectasia and Rad3 related protein (ATR) inhibition. Expression of PAX3-FOXO1 in muscle progenitor cells is not only sufficient to increase sensitivity to ATR inhibition, but PAX3-FOXO1-expressing rhabdomyosarcoma cells also exhibit increased sensitivity to structurally diverse inhibitors of ATR. Mechanistically, ATR inhibition leads to replication stress exacerbation, decreased BRCA1 phosphorylation and reduced homologous recombination-mediated DNA repair pathway activity. Consequently, ATR inhibitor treatment increases sensitivity of ARMS cells to PARP1 inhibition in vitro, and combined treatment with ATR and PARP1 inhibitors induces complete regression of primary patient-derived ARMS xenografts in vivo. Lastly, a genome-wide CRISPR activation screen (CRISPRa) in combination with transcriptional analyses of ATR inhibitor resistant ARMS cells identifies the RAS-MAPK pathway and its targets, the FOS gene family, as inducers of resistance to ATR inhibition. Our findings provide a rationale for upcoming biomarker-driven clinical trials of ATR inhibitors in patients suffering from ARMS.
© 2022. The Author(s).
Conflict of interest statement
A.M.W. is employed by Bayer AG. A.G.H. has received research funding from Bayer AG. The remaining authors declare no competing interests.
Figures





References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

