Genome-wide bidirectional CRISPR screens identify mucins as host factors modulating SARS-CoV-2 infection
- PMID: 35879412
- PMCID: PMC9355872
- DOI: 10.1038/s41588-022-01131-x
Genome-wide bidirectional CRISPR screens identify mucins as host factors modulating SARS-CoV-2 infection
Abstract
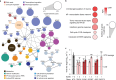
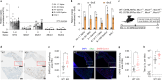
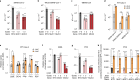
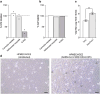
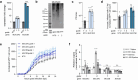
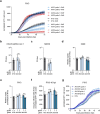
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) causes a range of symptoms in infected individuals, from mild respiratory illness to acute respiratory distress syndrome. A systematic understanding of host factors influencing viral infection is critical to elucidate SARS-CoV-2-host interactions and the progression of Coronavirus disease 2019 (COVID-19). Here, we conducted genome-wide CRISPR knockout and activation screens in human lung epithelial cells with endogenous expression of the SARS-CoV-2 entry factors ACE2 and TMPRSS2. We uncovered proviral and antiviral factors across highly interconnected host pathways, including clathrin transport, inflammatory signaling, cell-cycle regulation, and transcriptional and epigenetic regulation. We further identified mucins, a family of high molecular weight glycoproteins, as a prominent viral restriction network that inhibits SARS-CoV-2 infection in vitro and in murine models. These mucins also inhibit infection of diverse respiratory viruses. This functional landscape of SARS-CoV-2 host factors provides a physiologically relevant starting point for new host-directed therapeutics and highlights airway mucins as a host defense mechanism.
© 2022. The Author(s).
Conflict of interest statement
P.D.H. is a co-founder of Spotlight Therapeutics and Moment Biosciences and serves on the board of directors and scientific advisory boards, and is a scientific advisory board member to Vial Health and Serotiny. P.D.H. and S.K. are inventors on patents relating to CRISPR technologies. C.R.B. is a co-founder and Scientific Advisory Board member of Lycia Therapeutics, Palleon Pharmaceuticals, Enable Bioscience, Redwood Biosciences (a subsidiary of Catalent), OliLux Bio, Grace Science LLC and InterVenn Biosciences. Yale University (C.B.W) has a patent pending related to this work entitled ‘Compounds and Compositions for Treating, Ameliorating, and/or Preventing SARS-CoV-2 Infection and/or Complications Thereof’. The remaining authors declare no competing interests.
Figures








References
Publication types
MeSH terms
Substances
Grants and funding
- K08 AI163369/AI/NIAID NIH HHS/United States
- R21 AI156731/AI/NIAID NIH HHS/United States
- P01 HL110873/HL/NHLBI NIH HHS/United States
- R01 HL125280/HL/NHLBI NIH HHS/United States
- DP5 OD021369/OD/NIH HHS/United States
- UH3 HL123645/HL/NHLBI NIH HHS/United States
- R01 AI109022/AI/NIAID NIH HHS/United States
- K08 AI128043/AI/NIAID NIH HHS/United States
- R01 AI140186/AI/NIAID NIH HHS/United States
- P01 HL108808/HL/NHLBI NIH HHS/United States
- P30 DK065988/DK/NIDDK NIH HHS/United States
- R01 CA200423/CA/NCI NIH HHS/United States
- R01 HL136961/HL/NHLBI NIH HHS/United States
- T32 AI007502/AI/NIAID NIH HHS/United States
- R01 AI157253/AI/NIAID NIH HHS/United States
- HHMI/Howard Hughes Medical Institute/United States
- P30 CA016086/CA/NCI NIH HHS/United States
- R01 AI141970/AI/NIAID NIH HHS/United States
- F32 CA250324/CA/NCI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

