Bidirectional genome-wide CRISPR screens reveal host factors regulating SARS-CoV-2, MERS-CoV and seasonal HCoVs
- PMID: 35879413
- PMCID: PMC11627114
- DOI: 10.1038/s41588-022-01110-2
Bidirectional genome-wide CRISPR screens reveal host factors regulating SARS-CoV-2, MERS-CoV and seasonal HCoVs
Abstract
CRISPR knockout (KO) screens have identified host factors regulating severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) replication. Here, we conducted a meta-analysis of these screens, which showed a high level of cell-type specificity of the identified hits, highlighting the necessity of additional models to uncover the full landscape of host factors. Thus, we performed genome-wide KO and activation screens in Calu-3 lung cells and KO screens in Caco-2 colorectal cells, followed by secondary screens in four human cell lines. This revealed host-dependency factors, including AP1G1 adaptin and ATP8B1 flippase, as well as inhibitors, including mucins. Interestingly, some of the identified genes also modulate Middle East respiratory syndrome coronavirus (MERS-CoV) and seasonal human coronavirus (HCoV) (HCoV-NL63 and HCoV-229E) replication. Moreover, most genes had an impact on viral entry, with AP1G1 likely regulating TMPRSS2 activity at the plasma membrane. These results demonstrate the value of multiple cell models and perturbational modalities for understanding SARS-CoV-2 replication and provide a list of potential targets for therapeutic interventions.
© 2022. The Author(s), under exclusive licence to Springer Nature America, Inc.
Conflict of interest statement
Competing interests
J.G.D. consults for Agios, Microsoft Research, Phenomic AI, BioNTech, and Pfizer; J.G.D consults for and has equity in Tango Therapeutics. J.G.D.’s interests were reviewed and are managed by the Broad Institute in accordance with its conflict-of-interest policies. The other authors declare no competing interests.
Figures
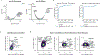






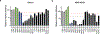
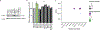
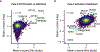








Update of
-
Bidirectional genome-wide CRISPR screens reveal host factors regulating SARS-CoV-2, MERS-CoV and seasonal coronaviruses.bioRxiv [Preprint]. 2021 May 21:2021.05.19.444823. doi: 10.1101/2021.05.19.444823. bioRxiv. 2021. Update in: Nat Genet. 2022 Aug;54(8):1090-1102. doi: 10.1038/s41588-022-01110-2. PMID: 34031654 Free PMC article. Updated. Preprint.
-
Bidirectional genome-wide CRISPR screens reveal host factors regulating SARS-CoV-2, MERS-CoV and seasonal HCoVs.Res Sq [Preprint]. 2021 May 27:rs.3.rs-555275. doi: 10.21203/rs.3.rs-555275/v1. Res Sq. 2021. Update in: Nat Genet. 2022 Aug;54(8):1090-1102. doi: 10.1038/s41588-022-01110-2. PMID: 34075371 Free PMC article. Updated. Preprint.
References
-
- Drosten C et al. Identification of a Novel Coronavirus in Patients with Severe Acute Respiratory Syndrome. N Engl J Med 348, 1967–1976 (2003). - PubMed
-
- Zaki AM, van Boheemen S, Bestebroer TM, Osterhaus ADME & Fouchier RAM Isolation of a Novel Coronavirus from a Man with Pneumonia in Saudi Arabia. N Engl J Med 367, 1814–1820 (2012). - PubMed
-
- van der Hoek L Human coronaviruses: what do they cause? Antivir. Ther. (Lond.) 12, 651–658 (2007). - PubMed
Methods-only references
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

