Cas13d knockdown of lung protease Ctsl prevents and treats SARS-CoV-2 infection
- PMID: 35879545
- PMCID: PMC10082993
- DOI: 10.1038/s41589-022-01094-4
Cas13d knockdown of lung protease Ctsl prevents and treats SARS-CoV-2 infection
Abstract
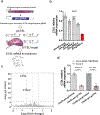
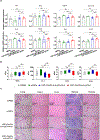
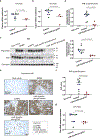
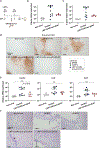
SARS-CoV-2 entry into cells requires specific host proteases; however, no successful in vivo applications of host protease inhibitors have yet been reported for treatment of SARS-CoV-2 pathogenesis. Here we describe a chemically engineered nanosystem encapsulating CRISPR-Cas13d, developed to specifically target lung protease cathepsin L (Ctsl) messenger RNA to block SARS-CoV-2 infection in mice. We show that this nanosystem decreases lung Ctsl expression in normal mice efficiently, specifically and safely. We further show that this approach extends survival of mice lethally infected with SARS-CoV-2, correlating with decreased lung virus burden, reduced expression of proinflammatory cytokines/chemokines and diminished severity of pulmonary interstitial inflammation. Postinfection treatment by this nanosystem dramatically lowers the lung virus burden and alleviates virus-induced pathological changes. Our results indicate that targeting lung protease mRNA by Cas13d nanosystem represents a unique strategy for controlling SARS-CoV-2 infection and demonstrate that CRISPR can be used as a potential treatment for SARS-CoV-2 infection.
© 2022. The Author(s), under exclusive licence to Springer Nature America, Inc.
Conflict of interest statement
Competing interests
Q.W., Z.C. and Y.D. are inventors on a patent (US Patent Application no. 17/626,482 entitled ‘Nanoparticle Systems for Targeted Delivery of CRISPR–Cas13 and Methods of Using Same’ filed 12 January, 2022, patent pending; European Patent Application no. 20840456.6 entitled ‘Nanoparticle Systems for Targeted Delivery of CRISPR–Cas13 and Methods of Using Same’ filed 12 January, 2022, patent pending) filed by Duke University that relates to the research reported in this paper. J.H. is a consultant for or owns shares in the following companies: Kingmed, MoreHealth, OptraScan, Genetron, Omnitura, Vetonco, York Biotechnology, Genecode, VIVA Biotech and Sisu Pharma and received grants from Zenith Epigenetics, BioXcel Therapeutics, Inc. and Fortis Therapeutics. The remaining authors declare no competing interests.
Figures











References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

