Na+ riboswitches regulate genes for diverse physiological processes in bacteria
- PMID: 35879547
- PMCID: PMC9337991
- DOI: 10.1038/s41589-022-01086-4
Na+ riboswitches regulate genes for diverse physiological processes in bacteria
Abstract
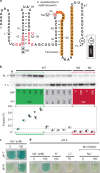
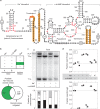
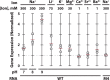
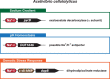
Organisms presumably have mechanisms to monitor and physiologically adapt to changes in cellular Na+ concentrations. Only a single bacterial protein has previously been demonstrated to selectively sense Na+ and regulate gene expression. Here we report a riboswitch class, previously called the 'DUF1646 motif', whose members selectively sense Na+ and regulate the expression of genes relevant to sodium biology. Many proteins encoded by Na+-riboswitch-regulated genes are annotated as metal ion transporters, whereas others are involved in mitigating osmotic stress or harnessing Na+ gradients for ATP production. Na+ riboswitches exhibit dissociation constants in the low mM range, and strongly reject all other alkali and alkaline earth ions. Likewise, only Na+ triggers riboswitch-mediated transcription and gene expression changes. These findings reveal that some bacteria use Na+ riboswitches to monitor, adjust and exploit Na+ concentrations and gradients, and in some instances collaborate with c-di-AMP riboswitches to coordinate gene expression during osmotic stress.
© 2022. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures






Comment in
-
A new class of metal-sensing RNA.Nat Chem Biol. 2022 Aug;18(8):798-799. doi: 10.1038/s41589-022-01087-3. Nat Chem Biol. 2022. PMID: 35879548 No abstract available.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

