A diagnostic classifier for gene expression-based identification of early Lyme disease
- PMID: 35879995
- PMCID: PMC9306241
- DOI: 10.1038/s43856-022-00127-2
A diagnostic classifier for gene expression-based identification of early Lyme disease
Abstract
Background: Lyme disease is a tick-borne illness that causes an estimated 476,000 infections annually in the United States. New diagnostic tests are urgently needed, as existing antibody-based assays lack sufficient sensitivity and specificity.
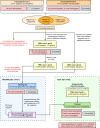
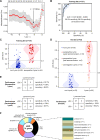
Methods: Here we perform transcriptome profiling by RNA sequencing (RNA-Seq), targeted RNA-Seq, and/or machine learning-based classification of 263 peripheral blood mononuclear cell samples from 218 subjects, including 94 early Lyme disease patients, 48 uninfected control subjects, and 57 patients with other infections (influenza, bacteremia, or tuberculosis). Differentially expressed genes among the 25,278 in the reference database are selected based on ≥1.5-fold change, ≤0.05 p value, and ≤0.001 false-discovery rate cutoffs. After gene selection using a k-nearest neighbor algorithm, the comparative performance of ten different classifier models is evaluated using machine learning.
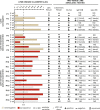
Results: We identify a 31-gene Lyme disease classifier (LDC) panel that can discriminate between early Lyme patients and controls, with 23 genes (74.2%) that have previously been described in association with clinical investigations of Lyme disease patients or in vitro cell culture and rodent studies of Borrelia burgdorferi infection. Evaluation of the LDC using an independent test set of samples from 63 subjects yields an overall sensitivity of 90.0%, specificity of 100%, and accuracy of 95.2%. The LDC test is positive in 85.7% of seronegative patients and found to persist for ≥3 weeks in 9 of 12 (75%) patients.
Conclusions: These results highlight the potential clinical utility of a gene expression classifier for diagnosis of early Lyme disease, including in patients negative by conventional serologic testing.
Keywords: Bacterial host response; Bacterial infection; Diagnostic markers; RNA sequencing; Targeted resequencing.
© The Author(s) 2022.
Conflict of interest statement
Competing interestsC.Y.C. and J.A. are on the scientific advisory board for the Bay Area Lyme Foundation. The other authors declare no competing interests.
Figures

References
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

