Restricted T-Cell Repertoire in the Epicardial Adipose Tissue of Non-ST Segment Elevation Myocardial Infarction Patients
- PMID: 35880176
- PMCID: PMC9307872
- DOI: 10.3389/fimmu.2022.845526
Restricted T-Cell Repertoire in the Epicardial Adipose Tissue of Non-ST Segment Elevation Myocardial Infarction Patients
Abstract
Aims: Human epicardial adipose tissue, a dynamic source of multiple bioactive factors, holds a close functional and anatomic relationship with the epicardial coronary arteries and communicates with the coronary artery wall through paracrine and vasocrine secretions. We explored the hypothesis that T-cell recruitment into epicardial adipose tissue (EAT) in patients with non-ST segment elevation myocardial infarction (NSTEMI) could be part of a specific antigen-driven response implicated in acute coronary syndrome onset and progression.
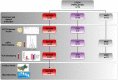
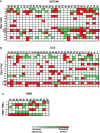
Methods and results: We enrolled 32 NSTEMI patients and 34 chronic coronary syndrome (CCS) patients undergoing coronary artery bypass grafting (CABG) and 12 mitral valve disease (MVD) patients undergoing surgery. We performed EAT proteome profiling on pooled specimens from three NSTEMI and three CCS patients. We performed T-cell receptor (TCR) spectratyping and CDR3 sequencing in EAT and peripheral blood mononuclear cells of 29 NSTEMI, 31 CCS, and 12 MVD patients. We then used computational modeling studies to predict interactions of the TCR beta chain variable region (TRBV) and explore sequence alignments. The EAT proteome profiling displayed a higher content of pro-inflammatory molecules (CD31, CHI3L1, CRP, EMPRINN, ENG, IL-17, IL-33, MMP-9, MPO, NGAL, RBP-4, RETN, VDB) in NSTEMI as compared to CCS (P < 0.0001). CDR3-beta spectratyping showed a TRBV21 enrichment in EAT of NSTEMI (12/29 patients; 41%) as compared with CCS (1/31 patients; 3%) and MVD (none) (ANOVA for trend P < 0.001). Of note, 11/12 (92%) NSTEMI patients with TRBV21 perturbation were at their first manifestation of ACS. Four patients with the first event shared a distinctive TRBV21-CDR3 sequence of 178 bp length and 2/4 were carriers of the human leukocyte antigen (HLA)-A*03:01 allele. A 3D analysis predicted the most likely epitope able to bind HLA-A3*01 and interact with the TRBV21-CDR3 sequence of 178 bp length, while the alignment results were consistent with microbial DNA sequences.
Conclusions: Our study revealed a unique immune signature of the epicardial adipose tissue, which led to a 3D modeling of the TCRBV/peptide/HLA-A3 complex, in acute coronary syndrome patients at their first event, paving the way for epitope-driven therapeutic strategies.
Keywords: NSTE ACS; T-cell receptor (TCR); antigen-driven immunity; computational modeling; epicardial adipose tissue (EAT); first acute myocardial infarction; immune response; precision medicine.
Copyright © 2022 Pedicino, Severino, Di Sante, De Rosa, Pirolli, Vinci, Pazzano, Giglio, Trotta, Russo, Ruggio, Pisano, d’Aiello, Canonico, Ciampi, Cianflone, Cianfanelli, Grimaldi, Filomia, Luciani, Glieca, Bruno, Massetti, Ria, Crea and Liuzzo.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures




References
-
- Maimaituxun G, Shimabukuro M, Fukuda D, Yagi S, Hirata Y, Iwase T, et al. Local Thickness of Epicardial Adipose Tissue Surrounding the Left Anterior Descending Artery Is a Simple Predictor of Coronary Artery Disease - New Prediction Model in Combination With Framingham Risk Score. Circ J (2018) 82:1369–78. doi: 10.1253/circj.CJ-17-1289 - DOI - PubMed
Publication types
MeSH terms
Substances
Associated data
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

