Harnessing the ubiquitin code to respond to environmental cues
- PMID: 35880291
- PMCID: PMC9400065
- DOI: 10.1042/EBC20210094
Harnessing the ubiquitin code to respond to environmental cues
Abstract
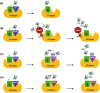
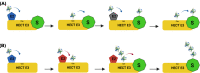
Ubiquitination is an essential post-translational signal that allows cells to adapt and respond to environmental stimuli. Substrate modifications range from a single ubiquitin molecule to complex polyubiquitin chains, where diverse chain topologies constitute a code that is utilized to modify the functions of proteins in numerous cellular signalling pathways. Diverse ubiquitin chain topologies are generated by linking the C-terminus of ubiquitin to one of seven lysine residues or the N-terminal methionine 1 residue of the preceding ubiquitin. Cooperative action between a large array of E2 conjugating and E3 ligase enzymes supports the formation of not only homotypic ubiquitin chains but also heterotypic mixed or branched chains. This complex array of chain topologies is recognized by proteins containing linkage-specific ubiquitin-binding domains and regulates numerous cellular pathways. Although many functions of the ubiquitin code in plants remain unknown, recent work suggests that specific chain topologies are associated with particular molecular processes. Deciphering the ubiquitin code and how plants utilize it to cope with the changing environment is essential to understand the regulatory mechanisms that underpin myriad stress responses and establishment of environmental tolerance.
Keywords: E2 enzyme; E3 ligase; chain topology; plant stress responses; proteasome; ubiquitin.
© 2022 The Author(s).
Conflict of interest statement
The authors declare that there are no competing interests associated with the manuscript.
Figures

Similar articles
-
Two different classes of E2 ubiquitin-conjugating enzymes are required for the mono-ubiquitination of proteins and elongation by polyubiquitin chains with a specific topology.Biochem J. 2008 Feb 1;409(3):723-9. doi: 10.1042/BJ20071338. Biochem J. 2008. PMID: 18042044
-
A ubiquitin ligase transfers preformed polyubiquitin chains from a conjugating enzyme to a substrate.Nature. 2007 Mar 15;446(7133):333-7. doi: 10.1038/nature05542. Epub 2007 Feb 18. Nature. 2007. PMID: 17310145
-
The K48-K63 Branched Ubiquitin Chain Regulates NF-κB Signaling.Mol Cell. 2016 Oct 20;64(2):251-266. doi: 10.1016/j.molcel.2016.09.014. Epub 2016 Oct 13. Mol Cell. 2016. PMID: 27746020
-
Post-translational regulation of ubiquitin signaling.J Cell Biol. 2019 Jun 3;218(6):1776-1786. doi: 10.1083/jcb.201902074. Epub 2019 Apr 18. J Cell Biol. 2019. PMID: 31000580 Free PMC article. Review.
-
Structural Diversity of Ubiquitin E3 Ligase.Molecules. 2021 Nov 4;26(21):6682. doi: 10.3390/molecules26216682. Molecules. 2021. PMID: 34771091 Free PMC article. Review.
Cited by
-
The Role of E3 Ubiquitin Ligase Gene FBK in Ubiquitination Modification of Protein and Its Potential Function in Plant Growth, Development, Secondary Metabolism, and Stress Response.Int J Mol Sci. 2025 Jan 19;26(2):821. doi: 10.3390/ijms26020821. Int J Mol Sci. 2025. PMID: 39859535 Free PMC article. Review.
-
Protein degrons and degradation: Exploring substrate recognition and pathway selection in plants.Plant Cell. 2024 Sep 3;36(9):3074-3098. doi: 10.1093/plcell/koae141. Plant Cell. 2024. PMID: 38701343 Free PMC article. Review.
-
The lowdown on breakdown: Open questions in plant proteolysis.Plant Cell. 2024 Sep 3;36(9):2931-2975. doi: 10.1093/plcell/koae193. Plant Cell. 2024. PMID: 38980154 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

