Nuclear Receptor NR1D1 Regulates Abdominal Aortic Aneurysm Development by Targeting the Mitochondrial Tricarboxylic Acid Cycle Enzyme Aconitase-2
- PMID: 35880522
- PMCID: PMC9674448
- DOI: 10.1161/CIRCULATIONAHA.121.057623
Nuclear Receptor NR1D1 Regulates Abdominal Aortic Aneurysm Development by Targeting the Mitochondrial Tricarboxylic Acid Cycle Enzyme Aconitase-2
Abstract
Background: Metabolic disorder increases the risk of abdominal aortic aneurysm (AAA). NRs (nuclear receptors) have been increasingly recognized as important regulators of cell metabolism. However, the role of NRs in AAA development remains largely unknown.
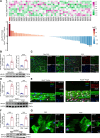
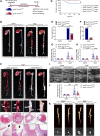
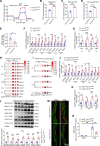
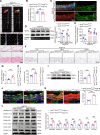
Methods: We analyzed the expression profile of the NR superfamily in AAA tissues and identified NR1D1 (NR subfamily 1 group D member 1) as the most highly upregulated NR in AAA tissues. To examine the role of NR1D1 in AAA formation, we used vascular smooth muscle cell (VSMC)-specific, endothelial cell-specific, and myeloid cell-specific conditional Nr1d1 knockout mice in both AngII (angiotensin II)- and CaPO4-induced AAA models.
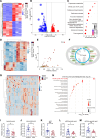
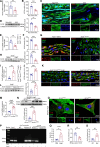
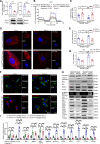
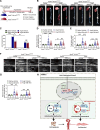
Results: Nr1d1 gene expression exhibited the highest fold change among all 49 NRs in AAA tissues, and NR1D1 protein was upregulated in both human and murine VSMCs from AAA tissues. The knockout of Nr1d1 in VSMCs but not endothelial cells and myeloid cells inhibited AAA formation in both AngII- and CaPO4-induced AAA models. Mechanistic studies identified ACO2 (aconitase-2), a key enzyme of the mitochondrial tricarboxylic acid cycle, as a direct target trans-repressed by NR1D1 that mediated the regulatory effects of NR1D1 on mitochondrial metabolism. NR1D1 deficiency restored the ACO2 dysregulation and mitochondrial dysfunction at the early stage of AngII infusion before AAA formation. Supplementation with αKG (α-ketoglutarate, a downstream metabolite of ACO2) was beneficial in preventing and treating AAA in mice in a manner that required NR1D1 in VSMCs.
Conclusions: Our data define a previously unrecognized role of nuclear receptor NR1D1 in AAA pathogenesis and an undescribed NR1D1-ACO2 axis involved in regulating mitochondrial metabolism in VSMCs. It is important that our findings suggest αKG supplementation as an effective therapeutic approach for AAA treatment.
Keywords: abdominal aortic aneurysm; alpha-Ketoglutarate; mitochondria; nuclear receptor; vascular smooth muscle.
Conflict of interest statement
None.
Figures
Comment in
-
Letter by Yang et al Regarding Article, "Nuclear Receptor NR1D1 Regulates Abdominal Aortic Aneurysm Development by Targeting the Mitochondrial Tricarboxylic Acid Cycle Enzyme Aconitase-2".Circulation. 2023 May 16;147(20):1560-1561. doi: 10.1161/CIRCULATIONAHA.122.063801. Epub 2023 May 15. Circulation. 2023. PMID: 37186681 No abstract available.
-
Response by Sun et al to Letter Regarding Article, "Nuclear Receptor NR1D1 Regulates Abdominal Aortic Aneurysm Development by Targeting the Mitochondrial Tricarboxylic Acid Cycle Enzyme Aconitase-2".Circulation. 2023 May 16;147(20):1562-1563. doi: 10.1161/CIRCULATIONAHA.123.064232. Epub 2023 May 15. Circulation. 2023. PMID: 37186684 No abstract available.
References
-
- Chaikof EL, Dalman RL, Eskandari MK, Jackson BM, Lee WA, Mansour MA, Mastracci TM, Mell M, Murad MH, Nguyen LL, et al. . The Society for Vascular Surgery practice guidelines on the care of patients with an abdominal aortic aneurysm. J Vasc Surg. 2018;67:2–77.e2. doi: 10.1016/j.jvs.2017.10.044 - PubMed
-
- Shi J, Yang Y, Cheng A, Xu G, He F. Metabolism of vascular smooth muscle cells in vascular diseases. Am J Physiol Heart Circ Physiol. 2020;319:H613–H631. doi: 10.1152/ajpheart.00220.2020 - PubMed
-
- Nasrallah CM, Horvath TL. Mitochondrial dynamics in the central regulation of metabolism. Nat Rev Endocrinol. 2014;10:650–658. doi: 10.1038/nrendo.2014.160 - PubMed
-
- Li Y, Ren P, Dawson A, Vasquez HG, Ageedi W, Zhang C, Luo W, Chen R, Li Y, Kim S, et al. . Single-cell transcriptome analysis reveals dynamic cell populations and differential gene expression patterns in control and aneurysmal human aortic tissue. Circulation. 2020;142:1374–1388. doi: 10.1161/CIRCULATIONAHA.120.046528 - PMC - PubMed
-
- Hollenberg AN. Metabolic health and nuclear-receptor sensitivity. N Engl J Med. 2012;366:1345–1347. doi: 10.1056/NEJMcibr1114529 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

