Donor Macrophages Modulate Rejection After Heart Transplantation
- PMID: 35880523
- PMCID: PMC9398940
- DOI: 10.1161/CIRCULATIONAHA.121.057400
Donor Macrophages Modulate Rejection After Heart Transplantation
Abstract
Background: Cellular rejection after heart transplantation imparts significant morbidity and mortality. Current immunosuppressive strategies are imperfect, target recipient T cells, and have adverse effects. The innate immune response plays an essential role in the recruitment and activation of T cells. Targeting the donor innate immune response would represent the earliest interventional opportunity within the immune response cascade. There is limited knowledge about donor immune cell types and functions in the setting of cardiac transplantation, and no current therapeutics exist for targeting these cell populations.
Methods: Using genetic lineage tracing, cell ablation, and conditional gene deletion, we examined donor mononuclear phagocyte diversity and macrophage function during acute cellular rejection of transplanted hearts in mice. We performed single-cell RNA sequencing on donor and recipient macrophages and monocytes at multiple time points after transplantation. On the basis of our imaging and single-cell RNA sequencing data, we evaluated the functional relevance of donor CCR2+ (C-C chemokine receptor 2) and CCR2- macrophages using selective cell ablation strategies in donor grafts before transplant. Last, we performed functional validation that donor macrophages signal through MYD88 (myeloid differentiation primary response protein 88) to facilitate cellular rejection.
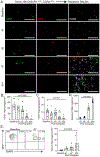
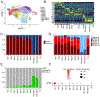
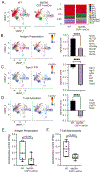
Results: Donor macrophages persisted in the rejecting transplanted heart and coexisted with recipient monocyte-derived macrophages. Single-cell RNA sequencing identified donor CCR2+ and CCR2- macrophage populations and revealed remarkable diversity among recipient monocytes, macrophages, and dendritic cells. Temporal analysis demonstrated that donor CCR2+ and CCR2- macrophages were transcriptionally distinct, underwent significant morphologic changes, and displayed unique activation signatures after transplantation. Although selective depletion of donor CCR2- macrophages reduced allograft survival, depletion of donor CCR2+ macrophages prolonged allograft survival. Pathway analysis revealed that donor CCR2+ macrophages are activated through MYD88/nuclear factor kappa light chain enhancer of activated B cells signaling. Deletion of MYD88 in donor macrophages resulted in reduced antigen-presenting cell recruitment, reduced ability of antigen-presenting cells to present antigen to T cells, decreased emergence of allograft-reactive T cells, and extended allograft survival.
Conclusions: Distinct populations of donor and recipient macrophages coexist within the transplanted heart. Donor CCR2+ macrophages are key mediators of allograft rejection, and deletion of MYD88 signaling in donor macrophages is sufficient to suppress rejection and extend allograft survival. This highlights the therapeutic potential of donor heart-based interventions.
Keywords: donor; heart; macrophages; sequence analysis, RNA; transplant.
Figures





Comment in
-
Donor macrophages influence rejection of transplanted hearts.Nat Rev Cardiol. 2022 Oct;19(10):641. doi: 10.1038/s41569-022-00764-z. Nat Rev Cardiol. 2022. PMID: 35974177 No abstract available.
-
Two-Faced Janus: CCR2-Expressing Macrophages and Their Dual Role in Allograft Rejection of the Transplanted Heart.Circulation. 2022 Aug 23;146(8):639-642. doi: 10.1161/CIRCULATIONAHA.122.061088. Epub 2022 Aug 22. Circulation. 2022. PMID: 35994560 No abstract available.
References
-
- Colvin M, Smith JM, Hadley N, Skeans MA, Carrico R, Uccellini K, Lehman R, Robinson A, Israni AK, Snyder JJ, et al. OPTN/SRTR 2016 Annual Data Report: Heart. Am J Transplant. 2018;18 Suppl 1:291–362. - PubMed
-
- Moayedi Y, Fan CPS, Cherikh WS, Stehlik J, Teuteberg JJ, Ross HJ and Khush KK. Survival Outcomes After Heart Transplantation: Does Recipient Sex Matter? Circ Heart Fail. 2019;12:e006218. - PubMed
-
- Chang DH, Kittleson MM and Kobashigawa JA. Immunosuppression following heart transplantation: prospects and challenges. Immunotherapy. 2014;6:181–94. - PubMed
-
- Alba AC, bain e, ng n, stein m, o’brien k, fortoutan f and ross h. Complications after Heart Transplantation: Hope for the Best, but Prepare for the Worst. International Journal of Transplantation Research and Medicine. 2016;2.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

