Transcript and metabolite network perturbations in lignin biosynthetic mutants of Arabidopsis
- PMID: 35880844
- PMCID: PMC9706439
- DOI: 10.1093/plphys/kiac344
Transcript and metabolite network perturbations in lignin biosynthetic mutants of Arabidopsis
Abstract
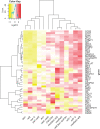
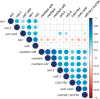
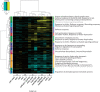
Lignin, one of the most abundant polymers in plants, is derived from the phenylpropanoid pathway, which also gives rise to an array of metabolites that are essential for plant fitness. Genetic engineering of lignification can cause drastic changes in transcription and metabolite accumulation with or without an accompanying development phenotype. To understand the impact of lignin perturbation, we analyzed transcriptome and metabolite data from the rapidly lignifying stem tissue in 13 selected phenylpropanoid mutants and wild-type Arabidopsis (Arabidopsis thaliana). Our dataset contains 20,974 expressed genes, of which over 26% had altered transcript levels in at least one mutant, and 18 targeted metabolites, all of which displayed altered accumulation in at least one mutant. We found that lignin biosynthesis and phenylalanine supply via the shikimate pathway are tightly co-regulated at the transcriptional level. The hierarchical clustering analysis of differentially expressed genes (DEGs) grouped the 13 mutants into 5 subgroups with similar profiles of mis-regulated genes. Functional analysis of the DEGs in these mutants and correlation between gene expression and metabolite accumulation revealed system-wide effects on transcripts involved in multiple biological processes.
© American Society of Plant Biologists 2022. All rights reserved. For permissions, please email: journals.permissions@oup.com.
Figures





Similar articles
-
A systems biology view of responses to lignin biosynthesis perturbations in Arabidopsis.Plant Cell. 2012 Sep;24(9):3506-29. doi: 10.1105/tpc.112.102574. Epub 2012 Sep 25. Plant Cell. 2012. PMID: 23012438 Free PMC article.
-
Indole Glucosinolate Biosynthesis Limits Phenylpropanoid Accumulation in Arabidopsis thaliana.Plant Cell. 2015 May;27(5):1529-46. doi: 10.1105/tpc.15.00127. Epub 2015 May 5. Plant Cell. 2015. PMID: 25944103 Free PMC article.
-
SG2-Type R2R3-MYB Transcription Factor MYB15 Controls Defense-Induced Lignification and Basal Immunity in Arabidopsis.Plant Cell. 2017 Aug;29(8):1907-1926. doi: 10.1105/tpc.16.00954. Epub 2017 Jul 21. Plant Cell. 2017. PMID: 28733420 Free PMC article.
-
Trends in lignin modification: a comprehensive analysis of the effects of genetic manipulations/mutations on lignification and vascular integrity.Phytochemistry. 2002 Oct;61(3):221-94. doi: 10.1016/s0031-9422(02)00211-x. Phytochemistry. 2002. PMID: 12359514 Review.
-
The cell biology of lignification in higher plants.Ann Bot. 2015 Jun;115(7):1053-74. doi: 10.1093/aob/mcv046. Epub 2015 Apr 15. Ann Bot. 2015. PMID: 25878140 Free PMC article. Review.
Cited by
-
Dissection of transcriptional events in graft incompatible reactions of "Bearss" lemon (Citrus limon) and "Valencia" sweet orange (C. sinensis) on a novel citrandarin (C. reticulata × Poncirus trifoliata) rootstock.Front Plant Sci. 2024 Jun 20;15:1421734. doi: 10.3389/fpls.2024.1421734. eCollection 2024. Front Plant Sci. 2024. PMID: 38966146 Free PMC article.
-
Identification of Genomic Regions Associated with Powdery Mildew Resistance in Watermelon through Genome-Wide Association Study.Plants (Basel). 2024 Sep 27;13(19):2708. doi: 10.3390/plants13192708. Plants (Basel). 2024. PMID: 39409578 Free PMC article.
-
Genome-wide association and selection studies for pod dehiscence resistance in the USDA soybean germplasm collection.PLoS One. 2025 Mar 28;20(3):e0318815. doi: 10.1371/journal.pone.0318815. eCollection 2025. PLoS One. 2025. PMID: 40153708 Free PMC article.
-
Pinoresinol rescues developmental phenotypes of Arabidopsis phenylpropanoid mutants overexpressing FERULATE 5-HYDROXYLASE.Proc Natl Acad Sci U S A. 2023 Aug;120(31):e2216543120. doi: 10.1073/pnas.2216543120. Epub 2023 Jul 24. Proc Natl Acad Sci U S A. 2023. PMID: 37487096 Free PMC article.
-
Suppression of the Arabidopsis cinnamoyl-CoA reductase 1-6 intronic T-DNA mutation by epigenetic modification.Plant Physiol. 2023 Aug 3;192(4):3001-3016. doi: 10.1093/plphys/kiad261. Plant Physiol. 2023. PMID: 37139862 Free PMC article.
References
-
- Anderson NA, Tobimatsu Y, Ciesielski PN, Ximenes E, Ralph J, Donohoe BS, Ladisch M, Chapple C (2015c) Manipulation of guaiacyl and syringyl monomer biosynthesis in an Arabidopsis cinnamyl alcohol dehydrogenase mutant results in atypical lignin biosynthesis and modified cell wall structure. Plant Cell 27: 2195–2209 - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

