Exon-based Phylogenomics and the Relationships of African Cichlid Fishes: Tackling the Challenges of Reconstructing Phylogenies with Repeated Rapid Radiations
- PMID: 35880863
- PMCID: PMC10198650
- DOI: 10.1093/sysbio/syac051
Exon-based Phylogenomics and the Relationships of African Cichlid Fishes: Tackling the Challenges of Reconstructing Phylogenies with Repeated Rapid Radiations
Abstract
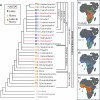
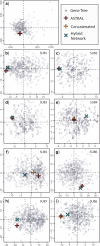
African cichlids (subfamily: Pseudocrenilabrinae) are among the most diverse vertebrates, and their propensity for repeated rapid radiation has made them a celebrated model system in evolutionary research. Nonetheless, despite numerous studies, phylogenetic uncertainty persists, and riverine lineages remain comparatively underrepresented in higher-level phylogenetic studies. Heterogeneous gene histories resulting from incomplete lineage sorting (ILS) and hybridization are likely sources of uncertainty, especially during episodes of rapid speciation. We investigate the relationships of Pseudocrenilabrinae and its close relatives while accounting for multiple sources of genetic discordance using species tree and hybrid network analyses with hundreds of single-copy exons. We improve sequence recovery for distant relatives, thereby extending the taxonomic reach of our probes, with a hybrid reference guided/de novo assembly approach. Our analyses provide robust hypotheses for most higher-level relationships and reveal widespread gene heterogeneity, including in riverine taxa. ILS and past hybridization are identified as the sources of genetic discordance in different lineages. Sampling of various Blenniiformes (formerly Ovalentaria) adds strong phylogenomic support for convict blennies (Pholidichthyidae) as sister to Cichlidae and points to other potentially useful protein-coding markers across the order. A reliable phylogeny with representatives from diverse environments will support ongoing taxonomic and comparative evolutionary research in the cichlid model system. [African cichlids; Blenniiformes; Gene tree heterogeneity; Hybrid assembly; Phylogenetic network; Pseudocrenilabrinae; Species tree.].
© The Author(s) 2022. Published by Oxford University Press on behalf of the Society of Systematic Biologists.
Figures


References
-
- Alter S., Munshi-South J., Stiassny M.L.J.. 2017. Genomewide SNP data reveal cryptic phylogeographic structure and microallopatric divergence in a rapids-adapted clade of cichlids from the Congo River. Mol. Ecol. 26:1401–1419. - PubMed
-
- Altschul S.F., Gish W., Miller W., Myers E.W., Lipman D.J.. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403–410. - PubMed
Publication types
MeSH terms
Associated data
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

