Mining of Thousands of Prokaryotic Genomes Reveals High Abundance of Prophages with a Strictly Narrow Host Range
- PMID: 35880895
- PMCID: PMC9426530
- DOI: 10.1128/msystems.00326-22
Mining of Thousands of Prokaryotic Genomes Reveals High Abundance of Prophages with a Strictly Narrow Host Range
Abstract
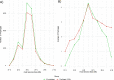
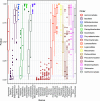
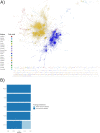
Phages and prophages are one of the principal modulators of microbial populations. However, much of their diversity is still poorly understood. Here, we extracted 33,624 prophages from 13,713 complete prokaryotic genomes to explore the prophage diversity and their relationships with their host. Our results reveal that prophages were present in 75% of the genomes studied. In addition, Enterobacterales were significantly enriched in prophages. We also found that pathogens are a significant reservoir of prophages. Finally, we determined that the prophage relatedness and the range of genomic hosts were delimited by the evolutionary relationships of their hosts. On a broader level, we got insights into the prophage population, identified in thousands of publicly available prokaryotic genomes, by comparing the prophage distribution and relatedness between them and their hosts. IMPORTANCE Phages and prophages play an essential role in controlling their host populations either by modulating the host abundance or providing them with genes that benefit the host. The constant growth in next-generation sequencing technology has caused the development of powerful computational tools to identify phages and prophages with high precision. Making it possible to explore the prophage populations integrated into host genomes on a large scale. However, it is still a new and under-explored area, and efforts are still required to identify prophage populations to understand their dynamics with their hosts.
Keywords: archaea; bacteria; bacteriophages; eubacteria; prophages; viral diversity.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

Similar articles
-
Host population structure and species resolution reveal prophage transmission dynamics.mBio. 2024 Oct 16;15(10):e0237724. doi: 10.1128/mbio.02377-24. Epub 2024 Sep 24. mBio. 2024. PMID: 39315801 Free PMC article.
-
Identification and characterization of Faecalibacterium prophages rich in diversity-generating retroelements.Microbiol Spectr. 2025 Feb 4;13(2):e0106624. doi: 10.1128/spectrum.01066-24. Epub 2024 Dec 31. Microbiol Spectr. 2025. PMID: 39745426 Free PMC article.
-
Prediction of Prophages and Their Host Ranges in Pathogenic and Commensal Neisseria Species.mSystems. 2022 Jun 28;7(3):e0008322. doi: 10.1128/msystems.00083-22. Epub 2022 Apr 14. mSystems. 2022. PMID: 35418239 Free PMC article.
-
Comparative genomics of phages and prophages in lactic acid bacteria.Antonie Van Leeuwenhoek. 2002 Aug;82(1-4):73-91. Antonie Van Leeuwenhoek. 2002. PMID: 12369206 Review.
-
Research progress of prophages.Yi Chuan. 2021 Mar 16;43(3):240-248. doi: 10.16288/j.yczz.20-355. Yi Chuan. 2021. PMID: 33724208 Review.
Cited by
-
Campylobacter prophage diversity reveals pervasive recombination between prophages from different Campylobacter species.Microbiol Spectr. 2024 Jan 11;12(1):e0279523. doi: 10.1128/spectrum.02795-23. Epub 2023 Dec 13. Microbiol Spectr. 2024. PMID: 38088548 Free PMC article.
-
Exploring Viral Interactions in Clavibacter Species: In Silico Analysis of Prophage Prevalence and Antiviral Defenses.Life (Basel). 2025 Jan 27;15(2):187. doi: 10.3390/life15020187. Life (Basel). 2025. PMID: 40003596 Free PMC article.
-
Lytic/Lysogenic Transition as a Life-History Switch.Virus Evol. 2024 Apr 3;10(1):veae028. doi: 10.1093/ve/veae028. eCollection 2024. Virus Evol. 2024. PMID: 38756985 Free PMC article.
-
Novel phages of Pseudomonas syringae unveil numerous potential auxiliary metabolic genes.J Gen Virol. 2024 Jun;105(6):001990. doi: 10.1099/jgv.0.001990. J Gen Virol. 2024. PMID: 38833289 Free PMC article.
-
Temperate phage-antibiotic synergy is widespread-extending to Pseudomonas-but varies by phage, host strain, and antibiotic pairing.mBio. 2025 Feb 5;16(2):e0255924. doi: 10.1128/mbio.02559-24. Epub 2024 Dec 20. mBio. 2025. PMID: 39704503 Free PMC article.
References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
