The molecular epidemiology of multiple zoonotic origins of SARS-CoV-2
- PMID: 35881005
- PMCID: PMC9348752
- DOI: 10.1126/science.abp8337
The molecular epidemiology of multiple zoonotic origins of SARS-CoV-2
Erratum in
-
Erratum for the Research Article "The molecular epidemiology of multiple zoonotic origins of SARS-CoV-2" by J. E. Pekar et al.Science. 2023 Oct 13;382(6667):eadl0585. doi: 10.1126/science.adl0585. Epub 2023 Oct 13. Science. 2023. PMID: 37824677 No abstract available.
Abstract
Understanding the circumstances that lead to pandemics is important for their prevention. We analyzed the genomic diversity of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) early in the coronavirus disease 2019 (COVID-19) pandemic. We show that SARS-CoV-2 genomic diversity before February 2020 likely comprised only two distinct viral lineages, denoted "A" and "B." Phylodynamic rooting methods, coupled with epidemic simulations, reveal that these lineages were the result of at least two separate cross-species transmission events into humans. The first zoonotic transmission likely involved lineage B viruses around 18 November 2019 (23 October to 8 December), and the separate introduction of lineage A likely occurred within weeks of this event. These findings indicate that it is unlikely that SARS-CoV-2 circulated widely in humans before November 2019 and define the narrow window between when SARS-CoV-2 first jumped into humans and when the first cases of COVID-19 were reported. As with other coronaviruses, SARS-CoV-2 emergence likely resulted from multiple zoonotic events.
Figures
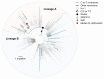
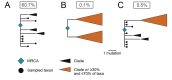
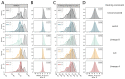

Comment in
-
Wildlife trade is likely the source of SARS-CoV-2.Science. 2022 Aug 26;377(6609):925-926. doi: 10.1126/science.add8384. Epub 2022 Aug 25. Science. 2022. PMID: 36007033
References
-
- Ren L.-L., Wang Y.-M., Wu Z.-Q., Xiang Z.-C., Guo L., Xu T., Jiang Y.-Z., Xiong Y., Li Y.-J., Li X.-W., Li H., Fan G.-H., Gu X.-Y., Xiao Y., Gao H., Xu J.-Y., Yang F., Wang X.-M., Wu C., Chen L., Liu Y.-W., Liu B., Yang J., Wang X.-R., Dong J., Li L., Huang C.-L., Zhao J.-P., Hu Y., Cheng Z.-S., Liu L.-L., Qian Z.-H., Qin C., Jin Q., Cao B., Wang J.-W., Identification of a novel coronavirus causing severe pneumonia in human: A descriptive study. Chin. Med. J. (Engl.) 133, 1015–1024 (2020). 10.1097/CM9.0000000000000722 - DOI - PMC - PubMed
-
- H. Ritchie, E. Mathieu, L. Rodés-Guirao, C. Appel, C. Giattino, E. Ortiz-Ospina, J. Hasell, B. Macdonald, S. Beltekian, X. Roser, Coronavirus Pandemic (COVID-19). Our World in Data (2022); https://ourworldindata.org/covid-deaths.
-
- Wu F., Zhao S., Yu B., Chen Y.-M., Wang W., Song Z.-G., Hu Y., Tao Z.-W., Tian J.-H., Pei Y.-Y., Yuan M.-L., Zhang Y.-L., Dai F.-H., Liu Y., Wang Q.-M., Zheng J.-J., Xu L., Holmes E. C., Zhang Y.-Z., A new coronavirus associated with human respiratory disease in China. Nature 579, 265–269 (2020). 10.1038/s41586-020-2008-3 - DOI - PMC - PubMed
MeSH terms
Grants and funding
- U19 AI135995/AI/NIAID NIH HHS/United States
- R01 AI153044/AI/NIAID NIH HHS/United States
- R01 AI132223/AI/NIAID NIH HHS/United States
- U54 CA260581/CA/NCI NIH HHS/United States
- T15 LM011271/LM/NLM NIH HHS/United States
- T32 AI007244/AI/NIAID NIH HHS/United States
- WT_/Wellcome Trust/United Kingdom
- R01 AI132244/AI/NIAID NIH HHS/United States
- R01 AI135992/AI/NIAID NIH HHS/United States
- R01 AI136056/AI/NIAID NIH HHS/United States
- U19 AI142790/AI/NIAID NIH HHS/United States
- 75N93021C00015/AI/NIAID NIH HHS/United States
- UL1 TR002550/TR/NCATS NIH HHS/United States
- U01 AI151812/AI/NIAID NIH HHS/United States
- U54 HG007480/HG/NHGRI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

