Establishing Linkages Among DNA Damage, Mutagenesis, and Genetic Diseases
- PMID: 35881568
- PMCID: PMC10201539
- DOI: 10.1021/acs.chemrestox.2c00155
Establishing Linkages Among DNA Damage, Mutagenesis, and Genetic Diseases
Abstract
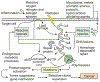
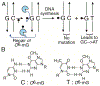
DNA damage by chemicals, radiation, or oxidative stress leads to a mutational spectrum, which is complex because it is determined in part by lesion structure, the DNA sequence context of the lesion, lesion repair kinetics, and the type of cells in which the lesion is replicated. Accumulation of mutations may give rise to genetic diseases such as cancer and therefore understanding the process underlying mutagenesis is of immense importance to preserve human health. Chemical or physical agents that cause cancer often leave their mutational fingerprints, which can be used to back-calculate the molecular events that led to disease. To make a clear link between DNA lesion structure and the mutations a given lesion induces, the field of single-lesion mutagenesis was developed. In the last three decades this area of research has seen much growth in several directions, which we attempt to describe in this Perspective.
Conflict of interest statement
The authors declare no competing financial interest.
Figures
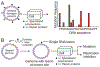
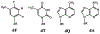




References
-
- Basu AK; Essigmann JM Site-specifically modified oligodeoxynucleotides as probes for the structural and biological effects of DNA-damaging agents. Chem. Res. Toxicol 1988, 1, 1–18. - PubMed
-
- Ohmori H; Friedberg EC; Fuchs RP; Goodman MF; Hanaoka F; Hinkle D; Kunkel TA; Lawrence CW; Livneh Z; Nohmi T; Prakash L; Prakash S; Todo T; Walker GC; Wang Z; Woodgate R The Y-family of DNA polymerases. Mol. Cell 2001, 8, 7–8. - PubMed
-
- Brenner S; Johnson M; Bridgham J; Golda G; Lloyd DH; Johnson D; Luo S; McCurdy S; Foy M; Ewan M; Roth R; George D; Eletr S; Albrecht G; Vermaas E; Williams SR; Moon K; Burcham T; Pallas M; DuBridge RB; Kirchner J; Fearon K; Mao J; Corcoran K Gene expression analysis by massively parallel signature sequencing (MPSS) on microbead arrays. Nat. Biotechnol 2000, i8, 630–634. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

