Reprogrammed Schwann Cells Organize into Dynamic Tracks that Promote Pancreatic Cancer Invasion
- PMID: 35881881
- PMCID: PMC9533012
- DOI: 10.1158/2159-8290.CD-21-1690
Reprogrammed Schwann Cells Organize into Dynamic Tracks that Promote Pancreatic Cancer Invasion
Abstract
Nerves are a component of the tumor microenvironment contributing to cancer progression, but the role of cells from nerves in facilitating cancer invasion remains poorly understood. Here we show that Schwann cells (SC) activated by cancer cells collectively function as tumor-activated Schwann cell tracks (TAST) that promote cancer cell migration and invasion. Nonmyelinating SCs form TASTs and have cell gene expression signatures that correlate with diminished survival in patients with pancreatic ductal adenocarcinoma. In TASTs, dynamic SCs form tracks that serve as cancer pathways and apply forces on cancer cells to enhance cancer motility. These SCs are activated by c-Jun, analogous to their reprogramming during nerve repair. This study reveals a mechanism of cancer cell invasion that co-opts a wound repair process and exploits the ability of SCs to collectively organize into tracks. These findings establish a novel paradigm of how cancer cells spread and reveal therapeutic opportunities.
Significance: How the tumor microenvironment participates in pancreatic cancer progression is not fully understood. Here, we show that SCs are activated by cancer cells and collectively organize into tracks that dynamically enable cancer invasion in a c-Jun-dependent manner. See related commentary by Amit and Maitra, p. 2240. This article is highlighted in the In This Issue feature, p. 2221.
©2022 The Authors; Published by the American Association for Cancer Research.
Figures


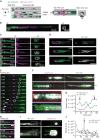
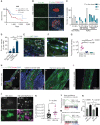
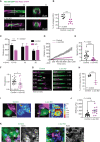

![Figure 7. c-Jun–reprogrammed SCs promote cancer invasion in vivo. A, Histologic analysis of injected murine sciatic nerves in P0-CRE− (WT) and P0-CRE+ (c-Jun KO) c-Junfl/fl mice. Representative samples of cancer invasion detected by H&E staining. Boxes show the invasion of areas away from the site of injection. Scale bars, 2,000 μm. Box scale bars, 500 μm. B, Quantification of distance of nerve invasion by Panc02 cells (WT n = 7, c-Jun KO n = 11, mean ± SEM) and KPC cells (WT n = 13, c-Jun KO n = 11, mean ± SEM). C, Representative images of murine hind limbs after Panc02 cancer cell injections show less paralysis in c-Jun KO SC mice. D, Quantification of the maximum width of hind limb paw in P0-CRE− (WT) versus P0-CRE+ (c-Jun KO) c-Junfl/fl mice 10 days after cancer injection (n = 8 mice/condition, mean ± SEM). E, Quantification of sciatic nerve function in P0-CRE− (WT) versus P0-CRE+ (c-Jun KO) c-Junfl/fl mice (n = 8 mice/condition, mean ± SEM). F, Effect of SP600125 on sciatic nerve invasion. Quantification of the length of nerve invasion in mice injected with Panc02 and SP600125 [JNK inhibitor (JNKi)] or Panc02 and DMSO (n = 11 mice/treatment, mean ± SEM). G, Representative H&E staining images of sciatic nerves injected with Panc02 and SP600125 versus Panc02 and DMSO. Top images are from proximal nerve regions at the spinal cord. No cancer is seen within the SP600125-injected nerve. Bottom images are from the site of injection, distal to the spinal cord. CC, cancer cells. Scale bars, 200 μm. H, Effect of SP600125 on flank tumor growth. Quantification of tumor volume in mice coinjected with Panc02 and SP600125 and with Panc02 and DMSO (n = 5 mice/treatment, mean ± SEM). NS, not significant. I, Survival analysis of WT and c-Jun KO mice injected with Panc02 in the sciatic nerve (WT n = 9, c-Jun KO = 11, P = 0.0003).](https://cdn.ncbi.nlm.nih.gov/pmc/blobs/e26a/9533012/cfbc03ef7a5c/2454fig7.gif)
Comment in
-
The Boring Schwann Cells: Tumor Me-TAST-asis along Nerves.Cancer Discov. 2022 Oct 5;12(10):2240-2243. doi: 10.1158/2159-8290.CD-22-0829. Cancer Discov. 2022. PMID: 36196575 Free PMC article.
References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

