Keratinocyte-derived defensins activate neutrophil-specific receptors Mrgpra2a/b to prevent skin dysbiosis and bacterial infection
- PMID: 35882236
- PMCID: PMC9474599
- DOI: 10.1016/j.immuni.2022.06.021
Keratinocyte-derived defensins activate neutrophil-specific receptors Mrgpra2a/b to prevent skin dysbiosis and bacterial infection
Abstract
Healthy skin maintains a diverse microbiome and a potent immune system to fight off infections. Here, we discovered that the epithelial-cell-derived antimicrobial peptides defensins activated orphan G-protein-coupled receptors (GPCRs) Mrgpra2a/b on neutrophils. This signaling axis was required for effective neutrophil-mediated skin immunity and microbiome homeostasis. We generated mutant mouse lines lacking the entire Defensin (Def) gene cluster in keratinocytes or Mrgpra2a/b. Def and Mrgpra2 mutant animals both exhibited skin dysbiosis, with reduced microbial diversity and expansion of Staphylococcus species. Defensins and Mrgpra2 were critical for combating S. aureus infections and the formation of neutrophil abscesses, a hallmark of antibacterial immunity. Activation of Mrgpra2 by defensin triggered neutrophil release of IL-1β and CXCL2 which are vital for proper amplification and propagation of the antibacterial immune response. This study demonstrated the importance of epithelial-neutrophil signaling via the defensin-Mrgpra2 axis in maintaining healthy skin ecology and promoting antibacterial host defense.
Keywords: CXCL2; GPCR; IL-1β; Mrgpr; antimicrobial peptide; defensin; innate immunity; neutrophil; skin dysbiosis; skin microbiome.
Copyright © 2022 Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests Xinzhong Dong is a co-founder and a scientific advisory board member of Escient Pharmaceuticals, a company focused on developing small molecules targeting MRGPRs. N.K.A. has received previous grant support from Pfizer and Boehringer Ingelheim and was a paid consultant for Janssen Pharmaceuticals. L.S.M. is currently a full-time paid employee of Janssen Research & Development, the pharmaceutical companies of Johnson & Johnson, and owns Johnson & Johnson stock. L.S.M. performed all work at his prior affiliation at Johns Hopkins University School of Medicine and he has received prior grant support from AstraZeneca, Pfizer, Boehringer Ingelheim, Regeneron Pharmaceuticals, and Moderna Therapeutics; was a paid consultant for Armirall and Janssen Research and Development; was on the scientific advisory board of Integrated Biotherapeutics; and is a shareholder of Noveome Biotherapeutics, which are all developing therapeutics against infections (including S. aureus and other pathogens) and/or inflammatory conditions. B.S.K. has served as a consultant for AbbVie, ABRAX Japan, Almirall, Cara Therapeutics, Maruho, Menlo Therapeutics, Pfizer, and Third Rock Ventures. He has also participated on the advisory board for Almirall, Boehringer Ingelheim, Cara Therapeutics, Kiniksa Pharmaceuticals, Menlo Therapeutics, Regeneron Pharmaceuticals, Sanofi Genzyme, and Trevi Therapeutics. He is also Founder, Chief Scientific Officer, and stockholder of Nuogen Pharma, Inc. He is stockholder of Locus Biosciences. C.V. is a paid consultant and stockholder of AstraZeneca.
Figures
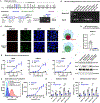


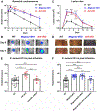
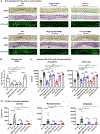


Comment in
-
The best offense is a good beta-defensin.Immunity. 2022 Sep 13;55(9):1586-1588. doi: 10.1016/j.immuni.2022.08.014. Immunity. 2022. PMID: 36103856
References
-
- Ahrens K, Schunck M, Podda GF, Meingassner J, Stuetz A, Schröder JM, Harder J, and Proksch E (2011). Mechanical and metabolic injury to the skin barrier leads to increased expression of murine β-defensin-1, −3, and −14. The Journal of investigative dermatology 131, 443–452. - PubMed
-
- Ali RS, Falconer A, Ikram M, Bissett CE, Cerio R, and Quinn AG (2001). Expression of the peptide antibiotics human beta defensin-1 and human beta defensin-2 in normal human skin. The Journal of investigative dermatology 117, 106–111. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 AR070116/AR/NIAMS NIH HHS/United States
- T32 GM136577/GM/NIGMS NIH HHS/United States
- HHMI/Howard Hughes Medical Institute/United States
- R01 AI146177/AI/NIAID NIH HHS/United States
- T32 AR074920/AR/NIAMS NIH HHS/United States
- R37 NS054791/NS/NINDS NIH HHS/United States
- R01 AR077007/AR/NIAMS NIH HHS/United States
- R01 AR073665/AR/NIAMS NIH HHS/United States
- R01 AR074846/AR/NIAMS NIH HHS/United States
- R21 AI167047/AI/NIAID NIH HHS/United States
- P30 AR079200/AR/NIAMS NIH HHS/United States
- R01 AR069502/AR/NIAMS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

