Lymph node homeostasis and adaptation to immune challenge resolved by fibroblast network mechanics
- PMID: 35882934
- PMCID: PMC9355877
- DOI: 10.1038/s41590-022-01272-5
Lymph node homeostasis and adaptation to immune challenge resolved by fibroblast network mechanics
Abstract
Emergent physical properties of tissues are not readily understood by reductionist studies of their constituent cells. Here, we show molecular signals controlling cellular, physical, and structural properties and collectively determine tissue mechanics of lymph nodes, an immunologically relevant adult tissue. Lymph nodes paradoxically maintain robust tissue architecture in homeostasis yet are continually poised for extensive expansion upon immune challenge. We find that in murine models of immune challenge, cytoskeletal mechanics of a cellular meshwork of fibroblasts determine tissue tension independently of extracellular matrix scaffolds. We determine that C-type lectin-like receptor 2 (CLEC-2)-podoplanin signaling regulates the cell surface mechanics of fibroblasts, providing a mechanically sensitive pathway to regulate lymph node remodeling. Perturbation of fibroblast mechanics through genetic deletion of podoplanin attenuates T cell activation. We find that increased tissue tension through the fibroblastic stromal meshwork is required to trigger the initiation of fibroblast proliferation and restore homeostatic cellular ratios and tissue structure through lymph node expansion.
© 2022. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures




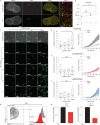







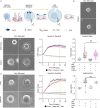


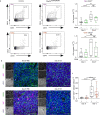
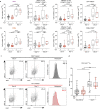

Comment in
-
Mechanics drive lymph node expansion.Nat Immunol. 2022 Aug;23(8):1139-1141. doi: 10.1038/s41590-022-01277-0. Nat Immunol. 2022. PMID: 35882935 No abstract available.
References
Publication types
MeSH terms
Substances
Grants and funding
- MR/T031646/1/MRC_/Medical Research Council/United Kingdom
- 19763/CRUK_/Cancer Research UK/United Kingdom
- MR/L009056//MRC_/Medical Research Council/United Kingdom
- MC_UU_00012/5/MRC_/Medical Research Council/United Kingdom
- CRUK-A19763/CRUK_/Cancer Research UK/United Kingdom
- MC_UU_12018/5/MRC_/Medical Research Council/United Kingdom
- MR/L009056/1/MRC_/Medical Research Council/United Kingdom
- MC-U12266B/MRC_/Medical Research Council/United Kingdom
- MC_UP_1205/1/MRC_/Medical Research Council/United Kingdom
- MR/W027437/1/MRC_/Medical Research Council/United Kingdom
- A19763/CRUK_/Cancer Research UK/United Kingdom
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

