BMP Signaling Pathway in Dentin Development and Diseases
- PMID: 35883659
- PMCID: PMC9317121
- DOI: 10.3390/cells11142216
BMP Signaling Pathway in Dentin Development and Diseases
Abstract
BMP signaling plays an important role in dentin development. BMPs and antagonists regulate odontoblast differentiation and downstream gene expression via canonical Smad and non-canonical Smad signaling pathways. The interaction of BMPs with their receptors leads to the formation of complexes and the transduction of signals to the canonical Smad signaling pathway (for example, BMP ligands, receptors, and Smads) and the non-canonical Smad signaling pathway (for example, MAPKs, p38, Erk, JNK, and PI3K/Akt) to regulate dental mesenchymal stem cell/progenitor proliferation and differentiation during dentin development and homeostasis. Both the canonical Smad and non-canonical Smad signaling pathways converge at transcription factors, such as Dlx3, Osx, Runx2, and others, to promote the differentiation of dental pulp mesenchymal cells into odontoblasts and downregulated gene expressions, such as those of DSPP and DMP1. Dysregulated BMP signaling causes a number of tooth disorders in humans. Mutation or knockout of BMP signaling-associated genes in mice results in dentin defects which enable a better understanding of the BMP signaling networks underlying odontoblast differentiation and dentin formation. This review summarizes the recent advances in our understanding of BMP signaling in odontoblast differentiation and dentin formation. It includes discussion of the expression of BMPs, their receptors, and the implicated downstream genes during dentinogenesis. In addition, the structures of BMPs, BMP receptors, antagonists, and dysregulation of BMP signaling pathways associated with dentin defects are described.
Keywords: BMP receptors; Smads; bone morphogenetic proteins (BMPs); canonical Smad signaling; dentin; dentin defects; downstream genes; non-canonical Smad signaling; odontoblasts.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
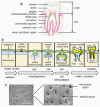
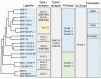

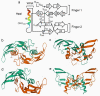

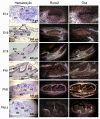


References
-
- Nanci A. Ten Cate’s Oral Histology: Development, Structure, and Function. 8th ed. Mosby; St. Louis, MI, USA: 2012. pp. 1–13.
-
- Chen S., Gluhak-Heinrich J., Martinez M., Li T., Wu Y., Chuang H.H., Chen L., Dong J., Gay I., MacDougall M. Bone morphogenetic protein 2 mediates dentin sialophosphoprotein expression and odontoblast differentiation via NF-Y signaling. J. Biol. Chem. 2008;283:19359–19370. doi: 10.1074/jbc.M709492200. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

