Next-Generation Sequencing Advances the Genetic Diagnosis of Cerebral Cavernous Malformation (CCM)
- PMID: 35883785
- PMCID: PMC9311989
- DOI: 10.3390/antiox11071294
Next-Generation Sequencing Advances the Genetic Diagnosis of Cerebral Cavernous Malformation (CCM)
Abstract
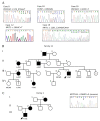
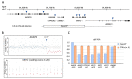
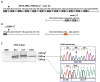
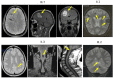
Cerebral Cavernous Malformation (CCM) is a cerebrovascular disease of genetic origin that predisposes to seizures, focal neurological deficits and fatal intracerebral hemorrhage. It may occur sporadically or in familial forms, segregating as an autosomal dominant condition with incomplete penetrance and highly variable expressivity. Its pathogenesis has been associated with loss-of-function mutations in three genes, namely KRIT1 (CCM1), CCM2 and PDCD10 (CCM3), which are implicated in defense mechanisms against oxidative stress and inflammation. Herein, we screened 21 Italian CCM cases using clinical exome sequencing and found six cases (~29%) with pathogenic variants in CCM genes, including a large 145−256 kb genomic deletion spanning the KRIT1 gene and flanking regions, and the KRIT1 c.1664C>T variant, which we demonstrated to activate a donor splice site in exon 16. The segregation of this cryptic splicing mutation was studied in a large Italian family (five affected and seven unaffected cases), and showed a largely heterogeneous clinical presentation, suggesting the implication of genetic modifiers. Moreover, by analyzing ad hoc gene panels, including a virtual panel of 23 cerebrovascular disease-related genes (Cerebro panel), we found two variants in NOTCH3 and PTEN genes, which could contribute to the abnormal oxidative stress and inflammatory responses to date implicated in CCM disease pathogenesis.
Keywords: KRIT1/CCM1; aberrant splicing; cerebral cavernous malformation (CCM); cerebrovascular disease; clinical exome sequencing (CES); genetic modifiers; next-generation sequencing (NGS).
Conflict of interest statement
The authors declare no conflict of interest.
Figures
References
-
- Fontanella M. Cerebral Cavernous Malformations (CCM) Minerva Medica; Rome, Italy: 2015.
-
- Rigamonti D. Cavernous Malformations of the Nervous System. Cambridge University Press; Cambridge, UK: 2011. p. 195.
-
- Flemming K.D. Incidence, Prevalence, and Clinical Presentation of Cerebral Cavernous Malformations. Methods Mol. Biol. 2020;2152:27–33. - PubMed
-
- Mabray M., Hart B. Clinical Imaging of Cerebral Cavernous Malformations: Computed Tomography and Magnetic Resonance Imaging. Methods Mol. Biol. 2020;2152:85–96. - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

