UFMylation System: An Emerging Player in Tumorigenesis
- PMID: 35884562
- PMCID: PMC9323365
- DOI: 10.3390/cancers14143501
UFMylation System: An Emerging Player in Tumorigenesis
Abstract
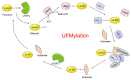
Ubiquitin-fold modifier 1 (UFM1), a newly identified ubiquitin-like molecule (UBLs), is evolutionarily expressed in multiple species except yeast. Similarly to ubiquitin, UFM1 is covalently attached to its substrates through a well-orchestrated three-step enzymatic reaction involving E1, the UFM1-activating enzyme (ubiquitin-like modifier-activating enzyme 5, UBA5); E2, the UFM1-conjugating enzyme 1 (UFC1); and E3, the UFM1-specific ligase 1 (UFL1). To date, numerous studies have shown that UFM1 modification is implicated in various cellular processes, including endoplasmic reticulum (ER) stress, DNA damage response and erythroid development. An abnormal UFM1 cascade is closely related to a variety of diseases, especially tumors. Herein, we summarize the process and functions of UFM1 modification, illustrating the relationship and mechanisms between aberrant UFMylation and diversified tumors, aiming to provide novel diagnostic biomarkers or therapeutic targets for cancer treatments.
Keywords: UFM1; UFMylation; post-translational modifications; tumorigenesis; ubiquitin-like molecules.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

Similar articles
-
UFMylation: A Unique & Fashionable Modification for Life.Genomics Proteomics Bioinformatics. 2016 Jun;14(3):140-146. doi: 10.1016/j.gpb.2016.04.001. Epub 2016 May 20. Genomics Proteomics Bioinformatics. 2016. PMID: 27212118 Free PMC article. Review.
-
Ufl1/RCAD, a Ufm1 E3 ligase, has an intricate connection with ER stress.Int J Biol Macromol. 2019 Aug 15;135:760-767. doi: 10.1016/j.ijbiomac.2019.05.170. Epub 2019 May 23. Int J Biol Macromol. 2019. PMID: 31129212 Review.
-
UFL1, a UFMylation E3 ligase, plays a crucial role in multiple cellular stress responses.Front Endocrinol (Lausanne). 2023 Feb 10;14:1123124. doi: 10.3389/fendo.2023.1123124. eCollection 2023. Front Endocrinol (Lausanne). 2023. PMID: 36843575 Free PMC article. Review.
-
Emerging role of protein modification by UFM1 in cancer.Biochem Biophys Res Commun. 2022 Dec 10;633:61-63. doi: 10.1016/j.bbrc.2022.08.093. Biochem Biophys Res Commun. 2022. PMID: 36344165 Review.
-
A novel type of E3 ligase for the Ufm1 conjugation system.J Biol Chem. 2010 Feb 19;285(8):5417-27. doi: 10.1074/jbc.M109.036814. Epub 2009 Dec 14. J Biol Chem. 2010. PMID: 20018847 Free PMC article.
Cited by
-
Systematic Analysis of UFMylation Family Genes in Tissues of Mice with Metabolic Dysfunction-Associated Steatotic Liver Disease.Genes (Basel). 2024 Dec 27;16(1):31. doi: 10.3390/genes16010031. Genes (Basel). 2024. PMID: 39858578 Free PMC article.
-
Role of UFMylation in tumorigenesis and cancer immunotherapy.Front Immunol. 2024 Aug 23;15:1454823. doi: 10.3389/fimmu.2024.1454823. eCollection 2024. Front Immunol. 2024. PMID: 39247188 Free PMC article. Review.
-
Emerging role of UFMylation in secretory cells involved in the endocrine system by maintaining ER proteostasis.Front Endocrinol (Lausanne). 2023 Jan 19;13:1085408. doi: 10.3389/fendo.2022.1085408. eCollection 2022. Front Endocrinol (Lausanne). 2023. PMID: 36743909 Free PMC article. Review.
-
A guide to UFMylation, an emerging posttranslational modification.FEBS J. 2023 Nov;290(21):5040-5056. doi: 10.1111/febs.16730. Epub 2023 Feb 8. FEBS J. 2023. PMID: 36680403 Free PMC article. Review.
-
The UFMylation pathway is impaired in Alzheimer's disease.bioRxiv [Preprint]. 2024 May 26:2024.05.24.595755. doi: 10.1101/2024.05.24.595755. bioRxiv. 2024. Update in: Mol Neurodegener. 2024 Dec 18;19(1):97. doi: 10.1186/s13024-024-00784-y. PMID: 38903110 Free PMC article. Updated. Preprint.
References
Publication types
Grants and funding
- 81670735/National Natural Science Foundation of China
- 81400802/National Natural Science Foundation of China
- 201701022/Clinical Research Project of Multi-Disciplinary Team, Shanghai Ninth People's Hospital, Shanghai JiaoTong University School of Medicine
- jyyq08201607/Outstanding Youth Training Project from Shanghai Ninth People's Hospital, Shanghai JiaoTong University School of Medicine
- 82000258/National Natural Science Foundation of China
LinkOut - more resources
Full Text Sources

