Differential Transcriptomic Profiles Following Stimulation with Lipopolysaccharide in Intestinal Organoids from Dogs with Inflammatory Bowel Disease and Intestinal Mast Cell Tumor
- PMID: 35884586
- PMCID: PMC9322748
- DOI: 10.3390/cancers14143525
Differential Transcriptomic Profiles Following Stimulation with Lipopolysaccharide in Intestinal Organoids from Dogs with Inflammatory Bowel Disease and Intestinal Mast Cell Tumor
Abstract
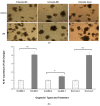
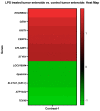
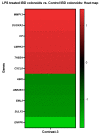
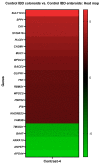
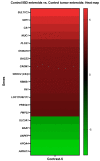
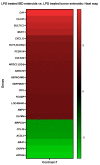
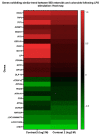
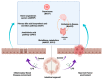
Lipopolysaccharide (LPS) is associated with chronic intestinal inflammation and promotes intestinal cancer progression in the gut. While the interplay between LPS and intestinal immune cells has been well-characterized, little is known about LPS and the intestinal epithelium interactions. In this study, we explored the differential effects of LPS on proliferation and the transcriptome in 3D enteroids/colonoids obtained from dogs with naturally occurring gastrointestinal (GI) diseases including inflammatory bowel disease (IBD) and intestinal mast cell tumor. The study objective was to analyze the LPS-induced modulation of signaling pathways involving the intestinal epithelia and contributing to colorectal cancer development in the context of an inflammatory (IBD) or a tumor microenvironment. While LPS incubation resulted in a pro-cancer gene expression pattern and stimulated proliferation of IBD enteroids and colonoids, downregulation of several cancer-associated genes such as Gpatch4, SLC7A1, ATP13A2, and TEX45 was also observed in tumor enteroids. Genes participating in porphyrin metabolism (CP), nucleocytoplasmic transport (EEF1A1), arachidonic acid, and glutathione metabolism (GPX1) exhibited a similar pattern of altered expression between IBD enteroids and IBD colonoids following LPS stimulation. In contrast, genes involved in anion transport, transcription and translation, apoptotic processes, and regulation of adaptive immune responses showed the opposite expression patterns between IBD enteroids and colonoids following LPS treatment. In brief, the crosstalk between LPS/TLR4 signal transduction pathway and several metabolic pathways such as primary bile acid biosynthesis and secretion, peroxisome, renin-angiotensin system, glutathione metabolism, and arachidonic acid pathways may be important in driving chronic intestinal inflammation and intestinal carcinogenesis.
Keywords: IBD; LPS; canine; colonoids; enteroids; mast cell tumor; microarray.
Conflict of interest statement
The authors declare no conflict of interest.
Figures




References
-
- Heinbockel L., Weindl G., Martinez-de-Tejada G., Correa W., Sanchez-Gomez S., Bárcena-Varela S., Goldmann T., Garidel P., Gutsmann T., Brandenburg K. Inhibition of Lipopolysaccharide- and Lipoprotein-Induced Inflammation by Antitoxin Peptide Pep19-2.5. Front. Immunol. 2018;9:1704. doi: 10.3389/fimmu.2018.01704. - DOI - PMC - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

