Tracking Clonal Evolution of Multiple Myeloma Using Targeted Next-Generation DNA Sequencing
- PMID: 35884979
- PMCID: PMC9313382
- DOI: 10.3390/biomedicines10071674
Tracking Clonal Evolution of Multiple Myeloma Using Targeted Next-Generation DNA Sequencing
Abstract
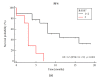
Clonal evolution drives treatment failure in multiple myeloma (MM). Here, we used a custom 372-gene panel to track genetic changes occurring during MM progression at different stages of the disease. A tumor-only targeted next-generation DNA sequencing was performed on 69 samples sequentially collected from 30 MM patients. The MAPK/ERK pathway was mostly affected with KRAS mutated in 47% of patients. Acquisition and loss of mutations were observed in 63% and 37% of patients, respectively. Four different patterns of mutation evolution were found: branching-, mutation acquisition-, mutation loss- and a stable mutational pathway. Better response to anti-myeloma therapy was more frequently observed in patients who followed the mutation loss-compared to the mutation acquisition pathway. More than two-thirds of patients had druggable genes mutated (including cases of heavily pre-treated disease). Only 7% of patients had a stable copy number variants profile. Consequently, a redistribution in stages according to R-ISS between the first and paired samples (R-ISS″) was seen. The higher the R-ISS″, the higher the risk of MM progression and death. We provided new insights into the genetics of MM evolution, especially in heavily pre-treated patients. Additionally, we confirmed that redefining R-ISS at MM relapse is of high clinical value.
Keywords: clonal evolution; multiple myeloma; next-generation sequencing.
Conflict of interest statement
The authors declare no conflict of interest.
Figures





References
-
- Melchor L., Brioli A., Wardell C.P., Murison A., Potter N.E., Kaiser M.F., Fryer R.A., Johnson D.C., Begum D.B., Wilson S.H., et al. Single-Cell Genetic Analysis Reveals the Composition of Initiating Clones and Phylogenetic Patterns of Branching and Parallel Evolution in Myeloma. Leukemia. 2014;28:1705–1715. doi: 10.1038/leu.2014.13. - DOI - PubMed
-
- Weinhold N., Ashby C., Rasche L., Chavan S.S., Stein C., Stephens O.W., Tytarenko R., Bauer M.A., Meissner T., Deshpande S., et al. Clonal Selection and Double-Hit Events Involving Tumor Suppressor Genes Underlie Relapse in Myeloma. Blood. 2016;128:1735–1744. doi: 10.1182/blood-2016-06-723007. - DOI - PMC - PubMed
-
- Walker B.A., Boyle E.M., Wardell C.P., Murison A., Begum D.B., Dahir N.M., Proszek P.Z., Johnson D.C., Kaiser M.F., Melchor L., et al. Mutational Spectrum, Copy Number Changes, and Outcome: Results of a Sequencing Study of Patients with Newly Diagnosed Myeloma. J. Clin. Oncol. 2015;33:3911–3920. doi: 10.1200/JCO.2014.59.1503. - DOI - PMC - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

