The Fate of IgE Epitopes and Coeliac Toxic Motifs during Simulated Gastrointestinal Digestion of Pizza Base
- PMID: 35885243
- PMCID: PMC9318710
- DOI: 10.3390/foods11142000
The Fate of IgE Epitopes and Coeliac Toxic Motifs during Simulated Gastrointestinal Digestion of Pizza Base
Abstract
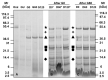
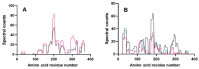
Understanding how food processing may modify allergen bioaccessibility and the evolution of immunologically active peptides in the gastrointestinal tract is essential if knowledge-based approaches to reducing the allergenicity of food are to be realised. A soy-enriched wheat-based pizza base was subjected to in vitro oral-gastro-duodenal digestion and resulting digests analysed using a combination of sodium dodecyl sulphate-polyacrylamide gel electrophoresis (SDS-PAGE) and mass spectrometry (MS). The digestion profile of pizza base resembled that of bread crust where higher temperatures during baking reduced protein solubility but still resulted in the generation of a complex mixture of peptides. MS profiling showed numerous peptides carrying IgE epitopes, and coeliac toxic motifs were in excess of 20-30 residues long and were only released after either 120 min of gastric digestion or a combination of gastric and duodenal digestion. In silico prediction tools showed an overestimated number of cleavage sites identified experimentally, with low levels of atypical peptic and chymotryptic cleavage sites identified particularly at glutamine residues. These data suggest that such alternative pepsin cleavage sites may play a role in digestion of glutamine-rich cereal foods. They also contribute to efforts to provide benchmarks for mapping in vitro digestion products of novel proteins which form part of the allergenicity risk assessment.
Keywords: IgE epitope; allergen; allergenicity risk assessment; coeliac toxic motif; gluten; in vitro digestion; mass spectrometry; soy; wheat.
Conflict of interest statement
The authors declare no conflict of interest in relation to the published work.
Figures





References
-
- Lyons S.A., Clausen M., Knulst A.C., Ballmer-Weber B.K., Fernandez-Rivas M., Barreales L., Bieli C., Dubakiene R., Fernandez-Perez C., Jedrzejczak-Czechowicz M., et al. Prevalence of Food Sensitization and Food Allergy in Children Across Europe. J. Allergy Clin. Immunol. Pract. 2020;8:2736–2746.e9. doi: 10.1016/j.jaip.2020.04.020. - DOI - PubMed
-
- Lyons S.A., Burney P.G.J., Ballmer-Weber B.K., Fernandez-Rivas M., Barreales L., Clausen M., Dubakiene R., Fernandez-Perez C., Fritsche P., Jedrzejczak-Czechowicz M., et al. Food Allergy in Adults: Substantial Variation in Prevalence and Causative Foods Across Europe. J. Allergy Clin. Immunol. Pract. 2019;7:1920–1928.e11. doi: 10.1016/j.jaip.2019.02.044. - DOI - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

