Immune-Related Genomic Schizophrenic Subtyping Identified in DLPFC Transcriptome
- PMID: 35885983
- PMCID: PMC9319783
- DOI: 10.3390/genes13071200
Immune-Related Genomic Schizophrenic Subtyping Identified in DLPFC Transcriptome
Abstract
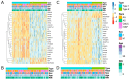
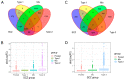
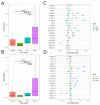
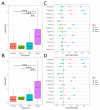
Well-documented evidence of the physiologic, genetic, and behavioral heterogeneity of schizophrenia suggests that diagnostic subtyping may clarify the underlying pathobiology of the disorder. Recent studies have demonstrated that increased inflammation may be a prominent feature of a subset of schizophrenics. However, these findings are inconsistent, possibly due to evaluating schizophrenics as a single group. In this study, we segregated schizophrenic patients into two groups ("Type 1", "Type 2") by their gene expression in the dorsolateral prefrontal cortex and explored biological differences between the subgroups. The study included post-mortem tissue samples that were sequenced in multiple, publicly available gene datasets using different sequencing methods. To evaluate the role of inflammation, the expression of genes in multiple components of neuroinflammation were examined: complement cascade activation, glial cell activation, pro-inflammatory mediator secretion, blood-brain barrier (BBB) breakdown, chemokine production and peripheral immune cell infiltration. The Type 2 schizophrenics showed widespread abnormal gene expression across all the neuroinflammation components that was not observed in Type 1 schizophrenics. Our results demonstrate the importance of separating schizophrenic patients into their molecularly defined subgroups and provide supporting evidence for the involvement of the immune-related pathways in a schizophrenic subset.
Keywords: inflammation; schizophrenia; subtypes; transcriptome.
Conflict of interest statement
The authors declare no conflict of interest.
Figures






References
-
- Chand G.B., Singhal P., Dwyer D.B., Wen J., Erus G., Doshi J., Srinivasan D., Mamourian E., Varol E., Hwang G., et al. Two schizophrenia imaging signatures and and their associations with cognition, psychopathology, and genetics in the general population. medRxiv. 2022 doi: 10.1101/2022.01.07.22268854. - DOI - PMC - PubMed

