AtGAP1 Promotes the Resistance to Pseudomonas syringae pv. tomato DC3000 by Regulating Cell-Wall Thickness and Stomatal Aperture in Arabidopsis
- PMID: 35886893
- PMCID: PMC9323218
- DOI: 10.3390/ijms23147540
AtGAP1 Promotes the Resistance to Pseudomonas syringae pv. tomato DC3000 by Regulating Cell-Wall Thickness and Stomatal Aperture in Arabidopsis
Abstract
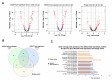
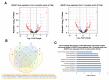
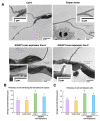
GTP is an important signaling molecule involved in the growth, development, and stress adaptability of plants. The functions are mediated via binding to GTPases which are in turn regulated by GTPase-activating proteins (GAPs). Satellite reports have suggested the positive roles of GAPs in regulating ABA signaling and pathogen resistance in plants. However, the molecular mechanisms that bring forth the pathogen resistance have remained unclear. In this study, we demonstrated that the expression of AtGAP1 was inducible by Pseudomonas syringae pv. tomato DC3000 (Pst DC3000). The overexpression of AtGAP1 in Arabidopsis promoted the expression of PR1 and the resistance to Pst DC3000. Proteomic analyses revealed the enhanced accumulation of cell-wall-modifying proteins as a result of AtGAP1 overexpression. By microscopic analyses, we showed that the overexpression of AtGAP1 resulted in increased thickness of the mesophyll cell wall and reduced stomatal aperture, which are effective strategies for restricting the entry of foliar pathogens. Altogether, we demonstrated that AtGAP1 increases the resistance to Pst DC3000 in Arabidopsis by promoting cellular strategies that restrict the entry of pathogens into the cells. These results point to a future direction for studying the modes of action of GAPs in regulating plant cell structures and disease resistance.
Keywords: GTP; GTPase activating protein (GAP); cell wall; pathogen resistance; stomatal aperture.
Conflict of interest statement
The authors declare no conflict of interest.
Figures




References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

