Characterization of Dense Granule Metalloproteinase INS-16 in Cryptosporidium parvum
- PMID: 35886965
- PMCID: PMC9315855
- DOI: 10.3390/ijms23147617
Characterization of Dense Granule Metalloproteinase INS-16 in Cryptosporidium parvum
Abstract
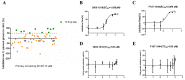
The protozoan pathogen Cryptosporidium parvum infects intestinal epithelial cells and causes diarrhea in humans and young animals. Among the more than 20 genes encoding insulinase-like metalloproteinases (INS), two are paralogs with high sequence identity. In this study, one of them, INS-16 encoded by the cgd3_4270 gene, was expressed and characterized in a comparative study of its sibling, INS-15 encoded by the cgd3_4260 gene. A full-length INS-16 protein and its active domain I were expressed in Escherichia coli, and antibodies against the domain I and an INS-16-specific peptide were produced in rabbits. In the analysis of the crude extract of oocysts, a ~60 kDa fragment of INS-16 rather than the full protein was recognized by polyclonal antibodies against the specific peptide, indicating that INS-16 undergoes proteolytic cleavage before maturation. The expression of the ins-16 gene peaked at the invasion phase of in vitro C. parvum culture, with the documented expression of the protein in both sporozoites and merozoites. Localization studies with antibodies showed significant differences in the distribution of the native INS-15 and INS-16 proteins in sporozoites and merozoites. INS-16 was identified as a dense granule protein in sporozoites and macrogamonts but was mostly expressed at the apical end of merozoites. We screened 48 candidate INS-16 inhibitors from the molecular docking of INS-16. Among them, two inhibited the growth of C. parvum in vitro (EC50 = 1.058 µM and 2.089 µM). The results of this study suggest that INS-16 may have important roles in the development of C. parvum and could be a valid target for the development of effective treatments.
Keywords: Cryptosporidium parvum; expression differences; invasion; metalloproteinase.
Conflict of interest statement
The authors declare no conflict of interest.
Figures




References
-
- Checkley W., White A.C., Jr., Jaganath D., Arrowood M.J., Chalmers R.M., Chen X.M., Fayer R., Griffiths J.K., Guerrant R.L., Hedstrom L., et al. A review of the global burden, novel diagnostics, therapeutics, and vaccine targets for Cryptosporidium. Lancet Infect. Dis. 2015;15:85–94. doi: 10.1016/S1473-3099(14)70772-8. - DOI - PMC - PubMed
-
- Kotloff K.L., Nataro J.P., Blackwelder W.C., Nasrin D., Farag T.H., Panchalingam S., Wu Y., Sow S.O., Sur D., Breiman R.F., et al. Burden and aetiology of diarrhoeal disease in infants and young children in developing countries (the Global Enteric Multicenter Study, GEMS): A prospective, case-control study. Lancet. 2013;382:209–222. doi: 10.1016/S0140-6736(13)60844-2. - DOI - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

