Nucleosome Assembly Protein 1, Nap1, Is Required for the Growth, Development, and Pathogenicity of Magnaporthe oryzae
- PMID: 35887015
- PMCID: PMC9316785
- DOI: 10.3390/ijms23147662
Nucleosome Assembly Protein 1, Nap1, Is Required for the Growth, Development, and Pathogenicity of Magnaporthe oryzae
Abstract
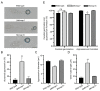
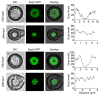
Magnaporthe oryzae is the causal agent of rice blast, leading to significant reductions in rice and wheat productivity. Nap1 is a conserved protein in eukaryotes involved in diverse physiological processes, such as nucleosome assembly, histone shuttling between the nucleus and cytoplasm, transcriptional regulation, and the cell cycle. Here, we identified Nap1 and characterized its roles in fungal development and virulence in M. oryzae. MoNap1 is involved in aerial hyphal and conidiophore differentiation, sporulation, appressorium formation, plant penetration, and virulence. ΔMonap1 generated a small, elongated, and malformed appressorium with an abnormally organized septin ring on hydrophobic surfaces. ΔMonap1 was more sensitive to cell wall integrity stresses but more resistant to microtubule stresses. MoNap1 interacted with histones H2A and H2B and the B-type cyclin (Cyc1). Moreover, a nuclear export signal (NES) domain is necessary for Nap1's roles in the regulation of the growth and pathogenicity of M. oryzae. In summary, NAP1 is essential for the growth, appressorium formation, and pathogenicity of M. oryzae.
Keywords: appressorium; cell wall integrity; histone; rice blast; septin ring; virulence.
Conflict of interest statement
The authors declare no conflict of interest.
Figures








References
-
- De Jong J.C., McCormack B.J., Smirnoff N., Talbot N.J. Glycerol generates turgor in rice blast. Nature. 1997;389:244–245. doi: 10.1038/38418. - DOI
MeSH terms
Substances
Supplementary concepts
Grants and funding
LinkOut - more resources
Full Text Sources
