Pre-Omicron Vaccine Breakthrough Infection Induces Superior Cross-Neutralization against SARS-CoV-2 Omicron BA.1 Compared to Infection Alone
- PMID: 35887023
- PMCID: PMC9320437
- DOI: 10.3390/ijms23147675
Pre-Omicron Vaccine Breakthrough Infection Induces Superior Cross-Neutralization against SARS-CoV-2 Omicron BA.1 Compared to Infection Alone
Abstract
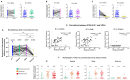
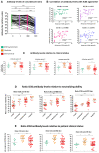
SARS-CoV-2 variants raise concern because of their high transmissibility and their ability to evade neutralizing antibodies elicited by prior infection or by vaccination. Here, we compared the neutralizing abilities of sera from 70 unvaccinated COVID-19 patients infected before the emergence of variants of concern (VOCs) and of 16 vaccine breakthrough infection (BTI) cases infected with Gamma or Delta against the ancestral B.1 strain, the Gamma, Delta and Omicron BA.1 VOCs using live virus. We further determined antibody levels against the Nucleocapsid (N) and full Spike proteins, the receptor-binding domain (RBD) and the N-terminal domain (NTD) of the Spike protein. Convalescent sera featured considerable variability in the neutralization of B.1 and in the cross-neutralization of different strains. Their neutralizing capacity moderately correlated with antibody levels against the Spike protein and the RBD. All but one convalescent serum failed to neutralize Omicron BA.1. Overall, convalescent sera from patients with moderate disease had higher antibody levels and displayed a higher neutralizing ability against all strains than patients with mild or severe forms of the disease. The sera from BTI cases fell into one of two categories: half the sera had a high neutralizing activity against the ancestral B.1 strain as well as against the infecting strain, while the other half had no or a very low neutralizing activity against all strains. Although antibody levels against the spike protein and the RBD were lower in BTI sera than in unvaccinated convalescent sera, most neutralizing sera also retained partial neutralizing activity against Omicron BA.1, suggestive of a better cross-neutralization and higher affinity of vaccine-elicited antibodies over virus-induced antibodies. Accordingly, the IC50: antibody level ratios were comparable for BTI and convalescent sera, but remained lower in the neutralizing convalescent sera from patients with moderate disease than in BTI sera. The neutralizing activity of BTI sera was strongly correlated with antibodies against the Spike protein and the RBD. Together, these findings highlight qualitative differences in antibody responses elicited by infection in vaccinated and unvaccinated individuals. They further indicate that breakthrough infection with a pre-Omicron variant boosts immunity and induces cross-neutralizing antibodies against different strains, including Omicron BA.1.
Keywords: Delta; Omicron BA.1; SARS-CoV-2; breakthrough infection; convalescent sera; neutralizing antibodies; variants of concern (VOCs).
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
-
- Korber B., Fischer W.M., Gnanakaran S., Yoon H., Theiler J., Abfalterer W., Hengartner N., Giorgi E.E., Bhattacharya T., Foley B., et al. Tracking Changes in SARS-CoV-2 Spike: Evidence that D614G Increases Infectivity of the COVID-19 Virus. Cell. 2020;182:812–827.e19. doi: 10.1016/j.cell.2020.06.043. - DOI - PMC - PubMed
-
- Lemieux J.E., Siddle K.J., Shaw B.M., Loreth C., Schaffner S.F., Gladden-Young A., Adams G., Fink T., Tomkins-Tinch C.H., Krasilnikova L.A., et al. Phylogenetic analysis of SARS-CoV-2 in Boston highlights the impact of superspreading events. Science. 2021;371:6529. doi: 10.1126/science.abe3261. - DOI - PMC - PubMed
-
- Wallace D.J., Ackland G.J. Abrupt increase in the UK coronavirus deathcase ratio in December 2020. medRxiv. 2021 doi: 10.1101/2021.01.21.21250264. - DOI
MeSH terms
Substances
Supplementary concepts
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

