The NEL Family of Bacterial E3 Ubiquitin Ligases
- PMID: 35887072
- PMCID: PMC9320238
- DOI: 10.3390/ijms23147725
The NEL Family of Bacterial E3 Ubiquitin Ligases
Abstract
Some pathogenic or symbiotic Gram-negative bacteria can manipulate the ubiquitination system of the eukaryotic host cell using a variety of strategies. Members of the genera Salmonella, Shigella, Sinorhizobium, and Ralstonia, among others, express E3 ubiquitin ligases that belong to the NEL family. These bacteria use type III secretion systems to translocate these proteins into host cells, where they will find their targets. In this review, we first introduce type III secretion systems and the ubiquitination process and consider the various ways bacteria use to alter the ubiquitin ligation machinery. We then focus on the members of the NEL family, their expression, translocation, and subcellular localization in the host cell, and we review what is known about the structure of these proteins, their function in virulence or symbiosis, and their specific targets.
Keywords: E3 ubiquitin ligases; Ralstonia; Salmonella; Shigella; Sinorhizobium; effectors; type III secretion.
Conflict of interest statement
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
Figures
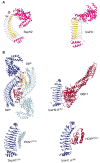

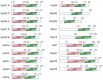
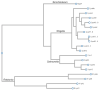
References
Publication types
MeSH terms
Substances
Grants and funding
- PID2019-106132RB-I00/AEI/10.13039/501100011033/Ministerio de Ciencia e Innovación - Agencia Estatal de Investigación
- P20_00576/Fondo Europeo de Desarrollo Regional (FEDER) y Consejería de Transformación Económica, Industria, Conocimiento y Universidades de la Junta de Andalucía
- US-1380805/Universidad de Sevilla, Fondo Europeo de Desarrollo Regional (FEDER) y Consejería de Transformación Económica, Industria, Conocimiento y Universidades de la Junta de Andalucía
LinkOut - more resources
Full Text Sources

