Unusual Association of NF-κB Components in Tumor-Associated Macrophages (TAMs) Promotes HSPG2-Mediated Immune-Escaping Mechanism in Breast Cancer
- PMID: 35887248
- PMCID: PMC9324337
- DOI: 10.3390/ijms23147902
Unusual Association of NF-κB Components in Tumor-Associated Macrophages (TAMs) Promotes HSPG2-Mediated Immune-Escaping Mechanism in Breast Cancer
Abstract
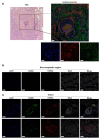
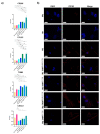
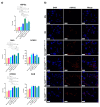
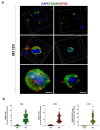
The cellular heterogeneity of the tumor environment of breast cancer (BC) is extremely complex and includes different actors such as neoplastic, stromal, and immunosuppressive cells, which contribute to the chemical and mechanical modification of the environment surrounding the tumor-exasperating immune-escaping mechanisms. In addition to molecular signals that make the tumor microenvironment (TME) unacceptable for the penetrance of the immune system, the physical properties of tumoral extracellular matrix (tECM) also have carved out a fundamental role in the processes of the protection of the tumor niche. Tumor-associated macrophages (TAMs), with an M2 immunosuppressive phenotype, are important determinants for the establishment of a tumor phenotype excluded from T cells. NF-κB transcription factors orchestrate innate immunity and represent the common thread between inflammation and cancer. Many studies have focused on canonical activation of NF-κB; however, activation of non-canonical signaling predicts poor survival and resistance to therapy. In this scenario, we demonstrated the existence of an unusual association of NF-κB components in TAMs that determines the deposition of HSPG2 that affects the stiffness of tECM. These results highlight a new mechanism counterbalanced between physical factors and a new perspective of mechano-pathology to be targeted to counteract immune evasion in BC.
Keywords: 3D culture; HSPG2; NF-κB; tumor-associated macrophages.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

References
-
- Liubomirski Y., Lerrer S., Meshel T., Rubinstein-Achiasaf L., Morein D., Wiemann S., Körner C., Ben-Baruch A. Tumor-Stroma-Inflammation Networks Promote Pro-Metastatic Chemokines and Aggressiveness Characteristics in Triple-Negative Breast Cancer. Front. Immunol. 2019;10:757. doi: 10.3389/fimmu.2019.00757. - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

