Sorting of Odor Dilutions Is a Meaningful Addition to Assessments of Olfactory Function as Suggested by Machine-Learning-Based Analyses
- PMID: 35887775
- PMCID: PMC9317381
- DOI: 10.3390/jcm11144012
Sorting of Odor Dilutions Is a Meaningful Addition to Assessments of Olfactory Function as Suggested by Machine-Learning-Based Analyses
Abstract
Background: The categorization of individuals as normosmic, hyposmic, or anosmic from test results of odor threshold, discrimination, and identification may provide a limited view of the sense of smell. The purpose of this study was to expand the clinical diagnostic repertoire by including additional tests.
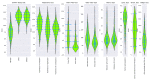
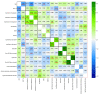
Methods: A random cohort of n = 135 individuals (83 women and 52 men, aged 21 to 94 years) was tested for odor threshold, discrimination, and identification, plus a distance test, in which the odor of peanut butter is perceived, a sorting task of odor dilutions for phenylethyl alcohol and eugenol, a discrimination test for odorant enantiomers, a lateralization test with eucalyptol, a threshold assessment after 10 min of exposure to phenylethyl alcohol, and a questionnaire on the importance of olfaction. Unsupervised methods were used to detect structure in the olfaction-related data, followed by supervised feature selection methods from statistics and machine learning to identify relevant variables.
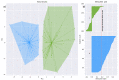
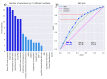
Results: The structure in the olfaction-related data divided the cohort into two distinct clusters with n = 80 and 55 subjects. Odor threshold, discrimination, and identification did not play a relevant role for cluster assignment, which, on the other hand, depended on performance in the two odor dilution sorting tasks, from which cluster assignment was possible with a median 100-fold cross-validated balanced accuracy of 77-88%.
Conclusions: The addition of an odor sorting task with the two proposed odor dilutions to the odor test battery expands the phenotype of olfaction and fits seamlessly into the sensory focus of standard test batteries.
Keywords: data science; machine learning; olfaction; olfactory testing; patients.
Conflict of interest statement
The authors declare no conflict of interest.
Figures




References
-
- Kobal G., Hummel T., Sekinger B., Barz S., Roscher S., Wolf S.R. “Sniffin’ Sticks”: Screening of olfactory performance. Rhinology. 1996;34:222–226. - PubMed
-
- Thomas-Danguin T., Rouby C., Sicard G., Vigouroux M., Farget V., Johanson A., Bengtzon A., Hall G., Ormel W., De Graaf C., et al. Development of the ETOC: A European test of olfactory capabilities. Rhinology. 2003;41:142–151. - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources

