Whole Exome Sequencing Enhanced Imputation Identifies 85 Metabolite Associations in the Alpine CHRIS Cohort
- PMID: 35888728
- PMCID: PMC9320943
- DOI: 10.3390/metabo12070604
Whole Exome Sequencing Enhanced Imputation Identifies 85 Metabolite Associations in the Alpine CHRIS Cohort
Abstract
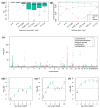
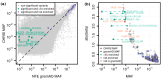
Metabolites are intermediates or end products of biochemical processes involved in both health and disease. Here, we take advantage of the well-characterized Cooperative Health Research in South Tyrol (CHRIS) study to perform an exome-wide association study (ExWAS) on absolute concentrations of 175 metabolites in 3294 individuals. To increase power, we imputed the identified variants into an additional 2211 genotyped individuals of CHRIS. In the resulting dataset of 5505 individuals, we identified 85 single-variant genetic associations, of which 39 have not been reported previously. Fifteen associations emerged at ten variants with >5-fold enrichment in CHRIS compared to non-Finnish Europeans reported in the gnomAD database. For example, the CHRIS-enriched ETFDH stop gain variant p.Trp286Ter (rs1235904433-hexanoylcarnitine) and the MCCC2 stop lost variant p.Ter564GlnextTer3 (rs751970792-carnitine) have been found in patients with glutaric acidemia type II and 3-methylcrotonylglycinuria, respectively, but the loci have not been associated with the respective metabolites in a genome-wide association study (GWAS) previously. We further identified three gene-trait associations, where multiple rare variants contribute to the signal. These results not only provide further evidence for previously described associations, but also describe novel genes and mechanisms for diseases and disease-related traits.
Keywords: ExWAS; GWAS; association study; imputation; metabolomics; whole-exome sequencing.
Conflict of interest statement
A.L. is currently employed by and holds stock in Regeneron Pharmaceuticals, though this work was initiated prior to employment and is unrelated. The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
Figures


References
-
- Hagenbeek F.A., Pool R., van Dongen J., Draisma H.H.M., Jan Hottenga J., Willemsen G., Abdellaoui A., Fedko I.O., den Braber A., Visser P.J., et al. Heritability estimates for 361 blood metabolites across 40 genome-wide association studies. Nat. Commun. 2020;11:39. doi: 10.1038/s41467-019-13770-6. - DOI - PMC - PubMed
-
- Hysi P.G., Mangino M., Christofidou P., Falchi M., Karoly E.D., NIHR Bioresource Investigators. Mohney R.P., Valdes A.M., Spector T.D., Menni C. Metabolome genome-wide association study identifies 74 novel genomic regions influencing plasma metabolites levels. Metabolites. 2022;12:61. doi: 10.3390/metabo12010061. - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Miscellaneous

