Comparative Genomic Analysis of the Marine Cyanobacterium Acaryochloris marina MBIC10699 Reveals the Impact of Phycobiliprotein Reacquisition and the Diversity of Acaryochloris Plasmids
- PMID: 35889093
- PMCID: PMC9324425
- DOI: 10.3390/microorganisms10071374
Comparative Genomic Analysis of the Marine Cyanobacterium Acaryochloris marina MBIC10699 Reveals the Impact of Phycobiliprotein Reacquisition and the Diversity of Acaryochloris Plasmids
Abstract
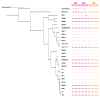
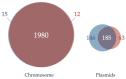
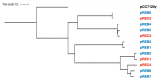
Acaryochloris is a marine cyanobacterium that synthesizes chlorophyll d, a unique chlorophyll that absorbs far-red lights. Acaryochloris is also characterized by the loss of phycobiliprotein (PBP), a photosynthetic antenna specific to cyanobacteria; however, only the type-strain A. marina MBIC11017 retains PBP, suggesting that PBP-related genes were reacquired through horizontal gene transfer (HGT). Acaryochloris is thought to have adapted to various environments through its huge genome size and the genes acquired through HGT; however, genomic information on Acaryochloris is limited. In this study, we report the complete genome sequence of A. marina MBIC10699, which was isolated from the same area of ocean as A. marina MBIC11017 as a PBP-less strain. The genome of A.marina MBIC10699 consists of a 6.4 Mb chromosome and four large plasmids totaling about 7.6 Mb, and the phylogenic analysis shows that A.marina MBIC10699 is the most closely related to A. marina MBIC11017 among the Acaryochloris species reported so far. Compared with A. marina MBIC11017, the chromosomal genes are highly conserved between them, while the genes encoded in the plasmids are significantly diverse. Comparing these genomes provides clues as to how the genes for PBPs were reacquired and what changes occurred in the genes for photosystems during evolution.
Keywords: Acaryochloris; comparative genome analysis; cyanobacteria.
Conflict of interest statement
The authors declare no conflict of interest.
Figures




References
-
- Miyashita H., Ikemoto H., Kurano N., Adachi K., Chihara M., Miyachi S. Chlorophyll d as a major pigment. Nature. 1996;383:402. doi: 10.1038/383402a0. - DOI
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

