Competitive Exclusion Bacterial Culture Derived from the Gut Microbiome of Nile Tilapia (Oreochromis niloticus) as a Resource to Efficiently Recover Probiotic Strains: Taxonomic, Genomic, and Functional Proof of Concept
- PMID: 35889095
- PMCID: PMC9321352
- DOI: 10.3390/microorganisms10071376
Competitive Exclusion Bacterial Culture Derived from the Gut Microbiome of Nile Tilapia (Oreochromis niloticus) as a Resource to Efficiently Recover Probiotic Strains: Taxonomic, Genomic, and Functional Proof of Concept
Abstract
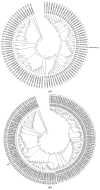
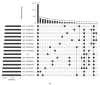
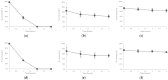
This study aims to mine a previously developed continuous-flow competitive exclusion culture (CFCEC) originating from the Tilapia gut microbiome as a rational and efficient autochthonous probiotic strain recovery source. Three isolated strains were tested on their adaptability to host gastrointestinal conditions, their antibacterial activities against aquaculture bacterial pathogens, and their antibiotic susceptibility patterns. Their genomes were fully sequenced, assembled, annotated, and relevant functions inferred, such as those related to pinpointed probiotic activities and phylogenomic comparative analyses to the closer reported strains/species relatives. The strains are possible candidates of novel genus/species taxa inside Lactococcus spp. and Priestia spp. (previously known as Bacillus spp.) These results were consistent with reports on strains inside these phyla exhibiting probiotic features, and the strains we found are expanding their known diversity. Furthermore, their pangenomes showed that these bacteria have indeed a set of so far uncharacterized genes that may play a role in the antagonism to competing strains or specific symbiotic adaptations to the fish host. In conclusion, CFCEC proved to effectively allow the enrichment and further pure culture isolation of strains with probiotic potential.
Keywords: Aeromonas hydrophila; Bacillus; Lactococcus lactis; Nile tilapia; Streptococcus agalactiae; continuos-flow competitive exclusion culture (CFCEC); freshwater fishes; microbiome; probiotics; whole-genome sequencing.
Conflict of interest statement
The authors declare no conflict of interest.
Figures






Similar articles
-
Unveiling the Probiotic Potential of the Anaerobic Bacterium Cetobacterium sp. nov. C33 for Enhancing Nile Tilapia (Oreochromis niloticus) Cultures.Microorganisms. 2023 Dec 5;11(12):2922. doi: 10.3390/microorganisms11122922. Microorganisms. 2023. PMID: 38138066 Free PMC article.
-
Establishment and characterization of a competitive exclusion bacterial culture derived from Nile tilapia (Oreochromis niloticus) gut microbiomes showing antibacterial activity against pathogenic Streptococcus agalactiae.PLoS One. 2019 May 3;14(5):e0215375. doi: 10.1371/journal.pone.0215375. eCollection 2019. PLoS One. 2019. PMID: 31050668 Free PMC article.
-
Evaluation of dietary single probiotic isolates and probiotic multistrain consortia in growth performance, gut histology, gut microbiota, immune regulation, and infection resistance of Nile tilapia, Oreochromis niloticus, shows superior monostrain performance.Fish Shellfish Immunol. 2023 Sep;140:108928. doi: 10.1016/j.fsi.2023.108928. Epub 2023 Jul 7. Fish Shellfish Immunol. 2023. PMID: 37423403
-
Probiotics in tilapia (Oreochromis niloticus) culture: Potential probiotic Lactococcus lactis culture conditions.J Biosci Bioeng. 2022 Mar;133(3):187-194. doi: 10.1016/j.jbiosc.2021.11.004. Epub 2021 Dec 15. J Biosci Bioeng. 2022. PMID: 34920949 Review.
-
Strategies for the Identification and Assessment of Bacterial Strains with Specific Probiotic Traits.Microorganisms. 2022 Jul 10;10(7):1389. doi: 10.3390/microorganisms10071389. Microorganisms. 2022. PMID: 35889107 Free PMC article. Review.
Cited by
-
Assessment of Encapsulated Probiotic Lactococcus lactis A12 Viability Using an In Vitro Digestion Model for Tilapia.Animals (Basel). 2024 Jul 5;14(13):1981. doi: 10.3390/ani14131981. Animals (Basel). 2024. PMID: 38998093 Free PMC article.
-
Improving the survival under gastric conditions of a potential multistrain probiotic produced in co-culture.AMB Express. 2025 Feb 6;15(1):20. doi: 10.1186/s13568-024-01810-4. AMB Express. 2025. PMID: 39915371 Free PMC article.
-
Unveiling the Probiotic Potential of the Anaerobic Bacterium Cetobacterium sp. nov. C33 for Enhancing Nile Tilapia (Oreochromis niloticus) Cultures.Microorganisms. 2023 Dec 5;11(12):2922. doi: 10.3390/microorganisms11122922. Microorganisms. 2023. PMID: 38138066 Free PMC article.
-
Design of an agro-industrial by-products-based media for the production of probiotic bacteria for fish nutrition.Sci Rep. 2024 Aug 2;14(1):17955. doi: 10.1038/s41598-024-68783-z. Sci Rep. 2024. PMID: 39095475 Free PMC article.
-
Interspecies Interactions within the Host: the Social Network of Group B Streptococcus.Infect Immun. 2023 Apr 18;91(4):e0044022. doi: 10.1128/iai.00440-22. Epub 2023 Mar 28. Infect Immun. 2023. PMID: 36975791 Free PMC article. Review.
References
-
- FAO . FAO Yearbook. Fishery and Aquaculture Statistics 2019/FAO Annuaire. Statistiques Des Pêches Et De L’aquaculture 2019/FAO Anuario. Estadísticas De Pesca Y Acuicultura 2019. FAO; Rome, Italy: 2021.
-
- Ullah A., Zuberi A., Ahmad M., Bashir Shah A., Younus N., Ullah S., Khattak M.N.K. Dietary administration of the commercially available probiotics enhanced the survival, growth, and innate immune responses in Mori (Cirrhinus mrigala) in a natural earthen polyculture system. Fish Shellfish Immunol. 2018;72:266–272. doi: 10.1016/j.fsi.2017.10.056. - DOI - PubMed
-
- Giri S., Sukumaran V., Sen S., Jena P. Effects of dietary supplementation of potential probiotic Bacillus subtilis VSG1 singularly or in combination with Lactobacillus plantarum VSG3 or/and Pseudomonas aeruginosa VSG2 on the growth, immunity and disease resistance of Labeo rohita. Aquac. Nutr. 2014;20:163–171. doi: 10.1111/anu.12062. - DOI
-
- Sankar H., Philip B., Philip R., Singh I. Effect of probiotics on digestive enzyme activities and growth of cichlids, Etroplus suratensis (Pearl spot) and Oreochromis mossambicus (Tilapia) Aquac. Nutr. 2017;23:852–864. doi: 10.1111/anu.12452. - DOI
-
- Aly S.M., Ahmed Y.A.-G., Ghareeb A.A.-A., Mohamed M.F. Studies on Bacillus subtilis and Lactobacillus acidophilus, as potential probiotics, on the immune response and resistance of Tilapia nilotica (Oreochromis niloticus) to challenge infections. Fish Shellfish Immunol. 2008;25:128–136. doi: 10.1016/j.fsi.2008.03.013. - DOI - PubMed
Grants and funding
- ING181-2016/Universidad de La Sabana
- INGPHD-6-2017/Universidad de La Sabana
- INGPHD-9-2019/Universidad de La Sabana
- 808-2018-contract CT 329-2019/MINCIENCIAS- Patrimonio Autónomo Fondo Nacional de Financiamiento para la Ciencia, la Tecnología y la Innovación Francisco José de Caldas
- 727-2015, contract CT 122-2017/MINCIENCIAS
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

