Role of Dissimilative Pathway of Komagataella phaffii (Pichia pastoris): Formaldehyde Toxicity and Energy Metabolism
- PMID: 35889185
- PMCID: PMC9321669
- DOI: 10.3390/microorganisms10071466
Role of Dissimilative Pathway of Komagataella phaffii (Pichia pastoris): Formaldehyde Toxicity and Energy Metabolism
Abstract
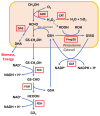
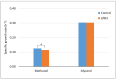
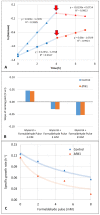
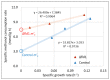
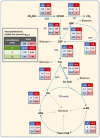
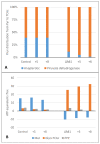
Komagataella phaffii (aka Pichia pastoris) is a yeast able to grow in methanol as the sole carbon and energy source. This substrate is converted into formaldehyde, a toxic intermediary that can either be assimilated to biomass or dissimilated to CO2 through the enzymes formaldehyde dehydrogenase (FLD) and formate dehydrogenase, also producing energy in the form of NADH. The dissimilative pathway has been described as an energy producing and a detoxifying route, but conclusive evidence has not been provided for this. In order to elucidate this theory, we generated mutants lacking the FLD activity (Δfld1) and used flux analysis to evaluate the metabolic impact of this disrupted pathway. Unexpectedly, we found that the specific growth rate of the Δfld1 strain was only slightly lower (92%) than the control. In contrast, the sensitivity to formaldehyde pulses (up to 8mM) was significantly higher in the Δfld1 mutant strain and was associated with a higher maintenance energy. In addition, the intracellular flux estimation revealed a high metabolic flexibility of K. phaffii in response to the disrupted pathway. Our results suggest that the role of the dissimilative pathway is mainly to protect the cells from the harmful effect of formaldehyde, as they were able to compensate for the energy provided from this pathway when disrupted.
Keywords: Komagataella phaffii; Pichia pastoris; dissimilative pathway; formaldehyde dehydrogenase; methanol.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
References
-
- Vanz A., Lünsdorf H., Adnan A., Nimtz M., Gurramkonda C., Khanna N., Rinas U. Physiological response of Pichia pastoris GS115 to methanol-induced high level production of the Hepatitis B surface antigen: Catabolic adaptation, stress responses, and autophagic processes. Microb. Cell Factories. 2012;11:103. doi: 10.1186/1475-2859-11-103. - DOI - PMC - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources

