Magnetic Levitation Patterns of Microfluidic-Generated Nanoparticle-Protein Complexes
- PMID: 35889600
- PMCID: PMC9324036
- DOI: 10.3390/nano12142376
Magnetic Levitation Patterns of Microfluidic-Generated Nanoparticle-Protein Complexes
Abstract
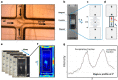
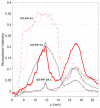
Magnetic levitation (MagLev) has recently emerged as a powerful method to develop diagnostic technologies based on the exploitation of the nanoparticle (NP)-protein corona. However, experimental procedures improving the robustness, reproducibility, and accuracy of this technology are largely unexplored. To contribute to filling this gap, here, we investigated the effect of total flow rate (TFR) and flow rate ratio (FRR) on the MagLev patterns of microfluidic-generated graphene oxide (GO)-protein complexes using bulk mixing of GO and human plasma (HP) as a reference. Levitating and precipitating fractions of GO-HP samples were characterized in terms of atomic force microscopy (AFM), bicinchoninic acid assay (BCA), and one-dimensional sodium dodecyl sulfate-polyacrylamide gel electrophoresis (1D SDS-PAGE), and nanoliquid chromatography-tandem mass spectrometry (nano-LC-MS/MS). We identified combinations of TFR and FRR (e.g., TFR = 35 μL/min and FRR (GO:HP) = 9:1 or TFR = 3.5 μL/min and FRR (GO:HP) = 19:1), leading to MagLev patterns dominated by levitating and precipitating fractions with bulk-like features. Since a typical MagLev experiment for disease detection is based on a sequence of optimization, exploration, and validation steps, this implies that the optimization (e.g., searching for optimal NP:HP ratios) and exploration (e.g., searching for MagLev signatures) steps can be performed using samples generated by bulk mixing. When these steps are completed, the validation step, which involves using human specimens that are often available in limited amounts, can be made by highly reproducible microfluidic mixing without any ex novo optimization process. The relevance of developing diagnostic technologies based on MagLev of coronated nanomaterials is also discussed.
Keywords: graphene oxide; magnetic levitation; microfluidics; protein corona.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

