SUMOylation and Viral Infections of the Brain
- PMID: 35890062
- PMCID: PMC9324588
- DOI: 10.3390/pathogens11070818
SUMOylation and Viral Infections of the Brain
Abstract
The small ubiquitin-like modifier (SUMO) system regulates numerous biological processes, including protein localization, stability and/or activity, transcription, and DNA repair. SUMO also plays critical roles in innate immunity and antiviral defense by mediating interferon (IFN) synthesis and signaling, as well as the expression and function of IFN-stimulated gene products. Viruses including human immunodeficiency virus-1, Zika virus, herpesviruses, and coronaviruses have evolved to exploit the host SUMOylation system to counteract the antiviral activities of SUMO proteins and to modify their own proteins for viral persistence and pathogenesis. Understanding the exploitation of SUMO is necessary for the development of effective antiviral therapies. This review summarizes the interplay between viruses and the host SUMOylation system, with a special emphasis on viruses with neuro-invasive properties that have pathogenic consequences on the central nervous system.
Keywords: HIV; SUMOylation; ZIKA; brain; coronavirus; cytomegalovirus; microglia; neuroinflammation; post-translational modifications.
Conflict of interest statement
The authors declare no competing interest.
Figures
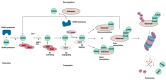

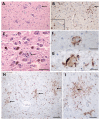
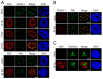

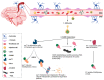
References
-
- Huber A.K., Irani D.N. Is the Concept of Central Nervous System Immune Privilege Irrelevant in the Setting of Acute Infection? [(accessed on 13 September 2020)];Front. Oncol. 2015 5 doi: 10.3389/fonc.2015.00099. Available online: https://www.frontiersin.org/articles/10.3389/fonc.2015.00099/full. - DOI - DOI - PMC - PubMed
-
- Boutell C., Cuchet-Lourenço D., Vanni E., Orr A., Glass M., McFarlane S., Everett R.D. A Viral Ubiquitin Ligase Has Substrate Preferential SUMO Targeted Ubiquitin Ligase Activity that Counteracts Intrinsic Antiviral Defence. [(accessed on 26 January 2021)];PLoS Pathog. 2011 7:e1002245. Available online: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3174244/ - PMC - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources

