Gene Losses and Plastome Degradation in the Hemiparasitic Species Plicosepalus acaciae and Plicosepalus curviflorus: Comparative Analyses and Phylogenetic Relationships among Santalales Members
- PMID: 35890506
- PMCID: PMC9317152
- DOI: 10.3390/plants11141869
Gene Losses and Plastome Degradation in the Hemiparasitic Species Plicosepalus acaciae and Plicosepalus curviflorus: Comparative Analyses and Phylogenetic Relationships among Santalales Members
Abstract
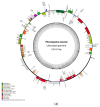
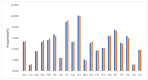
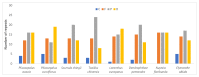
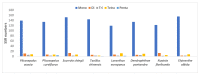
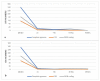
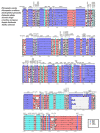
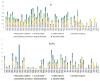
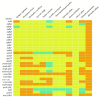
The Plicosepalus genus includes hemiparasitic mistletoe and belongs to the Loranthaceae family, and it has several medicinal uses. In the present study, we sequenced the complete plastomes of two species, Plicosepalus acaciae and Plicosepalus curviflorus, and compared them with the plastomes of photosynthetic species (hemiparasites) and nonphotosynthetic species (holoparasites) in the order Santalales. The complete chloroplast genomes of P. acaciae and P. curviflorus are circular molecules with lengths of 120,181 bp and 121,086 bp, respectively, containing 106 and 108 genes and 63 protein-coding genes, including 25 tRNA and 4 rRNA genes for each species. We observed a reduction in the genome size of P. acaciae and P. curviflorus and the loss of certain genes, although this reduction was less than that in the hemiparasite and holoparasitic cp genomes of the Santalales order. Phylogenetic analysis supported the taxonomic state of P. acaciae and P. curviflorus as members of the family Loranthaceae and tribe Lorantheae; however, the taxonomic status of certain tribes of Loranthaceae must be reconsidered and the species that belong to it must be verified. Furthermore, available chloroplast genome data of parasitic plants could help to strengthen efforts in weed management and encourage biotechnology research to improve host resistance.
Keywords: Plicosepalus acaciae; Plicosepalus curviflorus; comparative analysis; loranthaceae; mistletoe; phylogenetic relationship; plastome; plastome structure.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



References
-
- Liu B., Le C.T., Barrett R.L., Nickrent D.L., Chen Z., Lu L., Vidal-Russell R. Historical biogeography of Loranthaceae (Santalales): Diversification agrees with emergence of tropical forests and radiation of songbirds. Mol. Phylogenet. Evol. 2018;124:199–212. doi: 10.1016/j.ympev.2018.03.010. - DOI - PubMed
LinkOut - more resources
Full Text Sources
Miscellaneous

