A Newly Engineered A549 Cell Line Expressing ACE2 and TMPRSS2 Is Highly Permissive to SARS-CoV-2, Including the Delta and Omicron Variants
- PMID: 35891350
- PMCID: PMC9318744
- DOI: 10.3390/v14071369
A Newly Engineered A549 Cell Line Expressing ACE2 and TMPRSS2 Is Highly Permissive to SARS-CoV-2, Including the Delta and Omicron Variants
Abstract
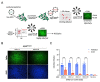
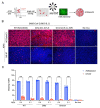
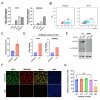
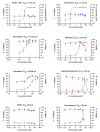
New variants of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) continue to emerge, causing surges, breakthrough infections, and devastating losses-underscoring the importance of identifying SARS-CoV-2 antivirals. A simple, accessible human cell culture model permissive to SARS-CoV-2 variants is critical for identifying and assessing antivirals in a high-throughput manner. Although human alveolar A549 cells are a valuable model for studying respiratory virus infections, they lack two essential host factors for SARS-CoV-2 infection: angiotensin-converting enzyme 2 (ACE2) and transmembrane serine protease 2 (TMPRSS2). SARS-CoV-2 uses the ACE2 receptor for viral entry and TMPRSS2 to prime the SARS-CoV-2 spike protein, both of which are negligibly expressed in A549 cells. Here, we report the generation of a suitable human cell line for SARS-CoV-2 studies by transducing human ACE2 and TMPRSS2 into A549 cells. We show that subclones highly expressing ACE2 and TMPRSS2 ("ACE2plus" and the subclone "ACE2plusC3") are susceptible to infection with SARS-CoV-2, including the delta and omicron variants. These subclones express more ACE2 and TMPRSS2 transcripts than existing commercial A549 cells engineered to express ACE2 and TMPRSS2. Additionally, the antiviral drugs EIDD-1931, remdesivir, nirmatrelvir, and nelfinavir strongly inhibit SARS-CoV-2 variants in our infection model. Our data show that ACE2plusC3 cells are highly permissive to SARS-CoV-2 infection and can be used to identify anti-SARS-CoV-2 drugs.
Keywords: A549; ACE2; EIDD-1931; SARS-CoV-2; TMPRSS2; delta and omicron variants; nirmatrelvir; remdesivir.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
Similar articles
-
SARS-CoV-2 Permissive glioblastoma cell line for high throughput antiviral screening.Antiviral Res. 2022 Jul;203:105342. doi: 10.1016/j.antiviral.2022.105342. Epub 2022 May 18. Antiviral Res. 2022. PMID: 35595082 Free PMC article.
-
Distinctive Roles of Furin and TMPRSS2 in SARS-CoV-2 Infectivity.J Virol. 2022 Apr 27;96(8):e0012822. doi: 10.1128/jvi.00128-22. Epub 2022 Mar 28. J Virol. 2022. PMID: 35343766 Free PMC article.
-
SARS-CoV-2 and SARS-CoV Spike-Mediated Cell-Cell Fusion Differ in Their Requirements for Receptor Expression and Proteolytic Activation.J Virol. 2021 Apr 12;95(9):e00002-21. doi: 10.1128/JVI.00002-21. Print 2021 Apr 12. J Virol. 2021. PMID: 33608407 Free PMC article.
-
Contributions of human ACE2 and TMPRSS2 in determining host-pathogen interaction of COVID-19.J Genet. 2021;100(1):12. doi: 10.1007/s12041-021-01262-w. J Genet. 2021. PMID: 33707363 Free PMC article. Review.
-
ACE2, TMPRSS2 distribution and extrapulmonary organ injury in patients with COVID-19.Biomed Pharmacother. 2020 Nov;131:110678. doi: 10.1016/j.biopha.2020.110678. Epub 2020 Aug 24. Biomed Pharmacother. 2020. PMID: 32861070 Free PMC article. Review.
Cited by
-
IFN-γ-mediated control of SARS-CoV-2 infection through nitric oxide.Front Immunol. 2023 Dec 15;14:1284148. doi: 10.3389/fimmu.2023.1284148. eCollection 2023. Front Immunol. 2023. PMID: 38162653 Free PMC article.
-
Cheminformatics-Based Discovery of Potential Chemical Probe Inhibitors of Omicron Spike Protein.Int J Mol Sci. 2022 Sep 7;23(18):10315. doi: 10.3390/ijms231810315. Int J Mol Sci. 2022. PMID: 36142242 Free PMC article.
-
Reciprocal enhancement of SARS-CoV-2 and influenza virus replication in human pluripotent stem cell-derived lung organoids1.Emerg Microbes Infect. 2023 Dec;12(1):2211685. doi: 10.1080/22221751.2023.2211685. Emerg Microbes Infect. 2023. PMID: 37161660 Free PMC article.
-
Efficient in vitro assay for evaluating drug efficacy and synergy against emerging SARS-CoV-2 strains.Antimicrob Agents Chemother. 2025 Feb 13;69(2):e0123324. doi: 10.1128/aac.01233-24. Epub 2024 Dec 17. Antimicrob Agents Chemother. 2025. PMID: 39688407 Free PMC article.
-
A multiplex method for rapidly identifying viral protease inhibitors.Mol Syst Biol. 2025 Feb;21(2):158-172. doi: 10.1038/s44320-024-00082-1. Epub 2025 Jan 6. Mol Syst Biol. 2025. PMID: 39762652 Free PMC article.
References
-
- Hoffmann M., Kleine-Weber H., Schroeder S., Krüger N., Herrler T., Erichsen S., Schiergens T.S., Herrler G., Wu N.-H., Nitsche A., et al. SARS-CoV-2 Cell Entry Depends on ACE2 and TMPRSS2 and Is Blocked by a Clinically Proven Protease Inhibitor. Cell. 2020;181:271–280.e278. doi: 10.1016/j.cell.2020.02.052. - DOI - PMC - PubMed
-
- Huang J., Hume A.J., Abo K.M., Werder R.B., Villacorta-Martin C., Alysandratos K.-D., Beermann M.L., Simone-Roach C., Lindstrom-Vautrin J., Olejnik J., et al. SARS-CoV-2 Infection of Pluripotent Stem Cell-Derived Human Lung Alveolar Type 2 Cells Elicits a Rapid Epithelial-Intrinsic Inflammatory Response. Cell Stem Cell. 2020;27:962–973.e967. doi: 10.1016/j.stem.2020.09.013. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Supplementary concepts
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

