Contrasting Epidemiology and Population Genetics of COVID-19 Infections Defined by Multilocus Genotypes in SARS-CoV-2 Genomes Sampled Globally
- PMID: 35891414
- PMCID: PMC9316073
- DOI: 10.3390/v14071434
Contrasting Epidemiology and Population Genetics of COVID-19 Infections Defined by Multilocus Genotypes in SARS-CoV-2 Genomes Sampled Globally
Abstract
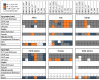
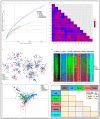
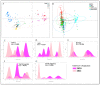
Since its emergence in 2019, SARS-CoV-2 has spread and evolved globally, with newly emerged variants of concern (VOCs) accounting for more than 500 million COVID-19 cases and 6 million deaths. Continuous surveillance utilizing simple genetic tools is needed to measure the viral epidemiological diversity, risk of infection, and distribution among different demographics in different geographical regions. To help address this need, we developed a proof-of-concept multilocus genotyping tool and demonstrated its utility to monitor viral populations sampled in 2020 and 2021 across six continents. We sampled globally 22,164 SARS-CoV-2 genomes from GISAID (inclusion criteria: available clinical and demographic data). They comprised two study populations, “2020 genomes” (N = 5959) sampled from December 2019 to September 2020 and “2021 genomes” (N = 16,205) sampled from 15 January to 15 March 2021. All genomes were aligned to the SARS-CoV-2 reference genome and amino acid polymorphisms were called with quality filtering. Thereafter, 74 codons (loci) in 14 genes including orf1ab polygene (N = 9), orf3a, orf8, nucleocapsid (N), matrix (M), and spike (S) met the 0.01 minimum allele frequency criteria and were selected to construct multilocus genotypes (MLGs) for the genomes. At these loci, 137 mutant/variant amino acids (alleles) were detected with eight VOC-defining variant alleles, including N KR203&204, orf1ab (I265, F3606, and L4715), orf3a H57, orf8 S84, and S G614, being predominant globally with > 35% prevalence. Their persistence and selection were associated with peaks in the viral transmission and COVID-19 incidence between 2020 and 2021. Epidemiologically, older patients (≥20 years) compared to younger patients (<20 years) had a higher risk of being infected with these variants, but this association was dependent on the continent of origin. In the global population, the discriminant analysis of principal components (DAPC) showed contrasting patterns of genetic clustering with three (Africa, Asia, and North America) and two (North and South America) continental clusters being observed for the 2020 and 2021 global populations, respectively. Within each continent, the MLG repertoires (range 40−199) sampled in 2020 and 2021 were genetically differentiated, with ≤4 MLGs per repertoire accounting for the majority of genomes sampled. These data suggested that the majority of SARS-CoV-2 infections in 2020 and 2021 were caused by genetically distinct variants that likely adapted to local populations. Indeed, four GISAID clade-defined VOCs - GRY (Alpha), GH (Beta), GR (Gamma), and G/GK (Delta variant) were differentiated by their MLG signatures, demonstrating the versatility of the MLG tool for variant identification. Results from this proof-of-concept multilocus genotyping demonstrates its utility for SARS-CoV-2 genomic surveillance and for monitoring its spatiotemporal epidemiology and evolution, particularly in response to control interventions including COVID-19 vaccines and chemotherapies.
Keywords: COVID-19; SARS-CoV-2; epidemiology; evolution; genetics; linkage; multilocus; mutation and transmission.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

References
-
- Talic S., Shah S., Wild H., Gasevic D., Maharaj A., Ademi Z., Li X., Xu W., Mesa-Eguiagaray I., Rostron J., et al. Effectiveness of public health measures in reducing the incidence of covid-19, SARS-CoV-2 transmission, and covid-19 mortality: Systematic review and meta-analysis. BMJ. 2021;375:e068302. doi: 10.1136/bmj-2021-068302. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Supplementary concepts
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

